Control region
Introduction
In the following, an alignment of nucleobases of the control region of selected species of the superorder Galloanserae is provided. Nucleobases that could be aligned with reasonable certainty are highlighted by various colours. Potential apomorphies are highlighted in red, while convergent nucleobase substitutions are highlighted in yellow.
Aligned partial control region sequences of thirty selected species of Galloanserae. Eleven unalignable regions (U-1 to U-11) and elven alignable regions (A-1 to A-11) are distinguished, with only the elongate terminal unalignable region (U-12) not being represented. [Colour symbols: dark blue for "G", light blue for "A", dark green for "T", light green for "C", pink for nucleotide transitions, purple for nucleotide transversions, orange for mixed transitions/ transversions, black for unresolved anseriform/galliform ground-pattern apomorphies, red for apomorphies in general, yellow for convergences, grey for deletions, and brown for insertions of varying lengths that are indicated by numbers]
As can be seen from the above sequences, the alignment is not always straightforward, and different regions vary with respect to alignability. Whenever multiple indels occur in variable regions, alignability is immediately lost. On the other hand, there are several conserved regions where only a limited number of nucleotide substitutions are encountered. A schematic map of the control region of Galloanserae is given below in order to provide an overview of the observed distribution of alignable and unalignable regions:
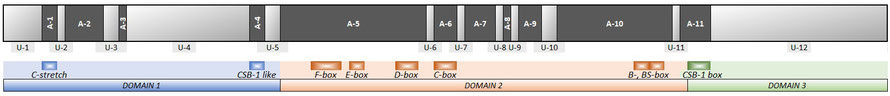
Schematic map of the control region in Galloanserae, with unalignable regions (U-1 to U-12) and alignable regions (A-1 to A-11) being distinguished. For comparison, the traditionally recognised domains and conserved boxes are presented in colour below. [Note that some of the alignable regions (e.g. A-2, A-7, A-8, A-9, as well as parts of A-5 and A-10) are lacking a corresponding traditional conserved box although no base substitutions were observed in Galloanserae]
Taken together, the control region exhibits a remarkable mix of variable and conserved regions. Because only the conserved regions are alignable, the control region turned out to be particularly informative in resolving phylogenetic relationships at the family level:
List of control-region apomorphies mapped onto the phylogeny of Galloanserae. While some relationships are particularly well resolved, others lack support altogether. Overall, the degree of homoplasy is limited, but some convergencies are encountered (summarised in the yellow box). Most parallelisms are easily recognised, because they are outnumbered by true apomorphies. Note that basal relationships within Galliformes (landfowl) and Anseriformes (waterfowl), respectively, are not shown, as there were no Struthioniformes or Neoaves included as outgroups.
One finding deserves particular consideration: from nuclear studies we know that
screamers (Anhimidae) represent the basalmost family of waterfowl, and that magpie-geese (Anseranatidae) diverge next (Prum et al., 2015; Kuhl et al., 2021). These results are corroborated by
morphological data (Livezey, 1986). Therefore, it is very surprising to find that control-region sequences provide strong evidence (by 11 apomorphies) for a sister-group relationship between
these two families. In other cases of mito-nuclear discordance, inconsistencies among different data sets are usually attributed to incomplete lineage sorting and/or introgression as a result of
hybridisation. However, because of the mere amount of conflicting data, in this particular case the hybridising species must have belonged to different families. Are viable hybrid offspring
really to be expected under these conditions?
References/Literature
Abbott CL, Double MC, Trueman JWH, Robinson A, and Cockburn A (2005), An unusual source of apparent mitochondrial heteroplasmy: duplicate mitochondrial control regions in Thalassarche albatrosses, Mol. Ecol. 14, 3605-13. (abstract)
Adawaren EO, Du Plessis M, Suleman E, Kindler D, Oosthuizen AO, Mukandiwa L, and Naidoo V (2020), The complete mitochondrial genome of Gyps coprotheres (Aves, Accipitridae, Accipitriformes): phylogenetic analysis of mitogenome among raptors, PeerJ 8, e:10034. (pdf)
Akiyama T, Nishida C, Momose K, Onuma M, Takami K, and masuda R (2017), Gene duplication and concerted evolution of mitochondrial DNA in crane species, Mol. Phylogenet. Evol. 106, 158-163. (abstract)
Al-Arab M, Höner zu Siederdissen C, Tout K, and Sahyoun AH (2017), Accurate annotation of protein-coding genes in mitochondrial genomes, Mol. Phylogenet. Evol. 106, 209-216. (abstract)
Aleix-Mata G, Ruiz-Ruano FJ, Perez JM, Sarasa M, and Sanchez A (2019), Complete mitochondrial genomes of the Western Caperacillie Tetrao urogallus (Phasianidae, Tetraoninae), Zootaxa 4550, 585-593. (pdf)
Alexeyev M (2020), Mitochondrial DNA: the common confusions, Mitochondrial DNA A 31, 45-47. (pdf)
Alfieri JM, Johnson T, Linderholm A, Blackmon H, and Athrey GN (2023), Genomic investigation refutes record of most diverged avian hybrid, Ecol. Evol. 13, e:9689. (pdf)
Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, and Sanger F (1981), Sequence and organization of the human mitochondrial genome, Nature 290, 457-465. (abstract)
Andreu-Sánchez S, Chen W, Stiller J, and Zhang G (2020), Multiple origins of a frame shift insertion in a mitochondrial gene in birds and turtles, GigaScience 10, 1-11. (free pdf)
Anmarkrud JA, and Lifjeld JT (2017), Complete mitochondrial genomes of eleven extinct or possibly extinct bird species, Mol. Ecol. Res. 17, 334-341. (abstract)
Baba Y, Fujimaki Y, Yoshii R, and Koike H (2001), Genetic variability in the mitochondrial control region of the Japanese Rock Ptarmigan Lagopus mutus japonicus, Jpn. J. Ornithol. 50, 53-64. (free pdf)
Baker AJ, and Marshall HD (1997), Mitochondrial control-region sequences as tools for understanding the evolution of avian taxa, In: “Avian molecular systematics and evolution” (Mindell DP, ed.), pp. 49-80, Academic Press, NY. (link)
Ballard JWO, and Whitlock MC (2004), The incomplete natural history of mitochondria, Mol. Ecol. 13, 729-744. (abstract)
Barker FK, Benesh MK, Vandergon AJ, and Lanyon Sam (2012), Contrasting evolutionary dynamics and information content of the avian mitochondrial control region and ND2 gene, PLOS ONE 7, e:46403. (pdf)
Barreira AS, Lijtmaer DA, and Tubaro PL (2016), The multiple applications of DNA barcodes in avian evolutionary studies, Genome 59, 899-911. (free pdf)
Barroso Lima NCB, and Prosdocimi F (2017), The heavy strand dilemma of vertebrate mitochondria on genome sequencing age: number of encoded genes or G+T content?, Mitochondrial DNA A 29, 300-302. (abstract)
Barroso Lima NCB (2018), Comparative mitogenomic analyses of Amazona parrots and Psittaciformes, Gen. Mol. Biol. 41, 593-604. (free pdf)
Bensch S, and Härlid A (2000), Mitochondrial genomic rearrangements in songbirds, Mol. Biol. Evol. 17, 107-113. (free pdf)
Berg T, Moum T, and Johansen S (1995), Variable numbers of simple tandem repeats make birds of the order Ciconiiformes heteroplasmic in their mitochondrial genomes, Curr. Genet. 27, 257-262. (abstract)
Bi D, Ding H, Wang Q, Jiang L, Lu W, Zhu R, Zeng J, Zhou S, Yang X, and Kan X (2019), Two new mitogenomes of Picidae (Aves, Piciformes): sequence, structure and phylogenetic analyses, Int. J. Biol. Macromol. 133, 683-692. (abstract)
Birks SM, and Edwards SV (2002), A phylogeny of the megapodes (Aves: Megapodiidae) based on nuclear and mitochondrial DNA sequences, Mol. Phylogenet. Evol. 23, 408-421. (abstract)
Brown GG, Gadeleta G, Pepe G, Saccone C, and Sbisá E (1986), Structural conservation and variation in the D-loop containing region of vertebrate mitochondrial DNA, J. Mol. Biol. 192, 503-511. (abstract)
Bruxaux J, Gabrielli M, Ashari H, Prys-Jones R, Joseph L, Milá B, Besnard G, and Thébaud C (2018), Recovering the evolutionary history of crowned pigeons (Columbidae: Goura): implications for the biogeography and conservation of New Guinean lowland birds, Mol. Phylogenet. Evol. 120, 248-258. (abstract)
Buckner JC, Ellingson R, Gold DA, Jones TL, and Jacobs DK (2018), Mitogenomics supports an unexpected taxonomic relationship for the extinct diving duck Chendytes lawi and definitely places the extinct Labrador Duck, Mol. Phylogenet. Evol. 122, 102-109. (abstract)
Buehler DM, and Baker AJ (2003), Characterization of the red knot (Calidris canutus) mitochondrial control region, Genome 46, 565-572. (abstract)
Cadahía L, Pinsker W, Negro JJ, Pavlicev M, Urios V, and Haring E (2009), Repeated sequence homogenisation between the control and pseudo-control regions in the mitochondrial genomes of the subfamily Aquilinae, J. Exp. Zool. 312B, 171-185. (abstract)
Camasamudram V, Fang JK, and Avadhani NG (2003), Transcription termination at the mouse mitochondrial H-strand promoter distal site requires an A/T rich sequence motiv and sequence specific DNA binding proteins, FEBS Journal 270, 1128-40. (pdf)
Campillo LC, Burns KJ, Moyle RG, and Manthey JD (2019), Mitochondrial genomes of the bird genus Piranga: rates of sequence evolution, and discordance between mitochondrial and nuclear markers, Mitochondrial DNA B 4, 2566-69. (free pdf)
Caparroz R, Rocha AV, Cabanne GS, Tubaro P, Aleixo A, Lemmon EM, and Lemmon AR (2018), Mitogenomes of two neotropical bird species and the multiple independent origin of mitochondrial gene orders in Passeriformes, Mol. Biol. Rep. 45, 279-285. (abstract)
Carson RJ, and Spicer GS (2003), A phylogenetic analysis of the emberizid sparrows based on three mitochondrial genes, Mol. Phylogenet. Evol. 29, 43-57. (abstract)
Carter JK, Kimball RT, Funk ER, Kane NC, Schield DR, Spellman GM, and Safran RJ (2023), Estimating phylogenies from genomes: a beginners review of commonly used genomic data in vertebrate phylogenomics, J. Hered., e: esac061. (free pdf)
Castresana J (2000), Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis, Mol. Biol. Evol. 17, 540-552. (free pdf)
Chen DS, Qian CJ, Ren QQ, Wang P, Yuan J, Jiang L, Bi D, Zhang Q, Wang Y, and Kan XZ (2015), Complete mitochondrial genome of the Chinese Hwamei Garrulax canorus (Aves: Passeriformes): the first representative of the Leiotrichidae family with a duplicated control region, Genet. Mol. Res. 14, 8964-76. (pdf)
Chen P, Han Y, Zhu C, Gao B, and Ruan L (2017), Complete mitochondrial genome of Porzana fusca and Porzana pusilla and phylogenetic relationship of 16 Rallidae species, Genetica 145, 559-573. (abstract)
Chen P, Huang Z, Zhu C, Han Y, Xu Z, Sun G, Zhang Z, Zhao D, Ge G, and Luzhang R (2020), Complete mitochondrial genome and phylogenetic analysis of Gruiformes and Charadriiformes, Pakistan J. Zool. 52, 425-439. (free pdf)
Chen W, Miao K, Wang J, Wang H, Sun W, Yuan S, Luo S, Zhu C, and Chang Q (2022), Five new mitogenomes sequences of Calidrine sandpipers (Aves: Charadriiformes) and comparative mitogenomics of genus Calidris, PeerJ 10, e:13268. (pdf)
Chen Y, Li F, Zhang Q, and Wang Q (2018), Complete mitochondrial genome of the Himalayan Monal Lophophorus impejanus (Phasianidae), with phylogenetic implication, Conservation Genet. Resour. 10, 877-880.(abstract)
Cho HJ, Eda M, Nishida S, Yasukochi Y, Chong JR, and Koike H (2009), Tandem duplication of mitochondrial DNA in the black-faced spoonbill, Platalea minor, Genes Genet. Syst. 84, 297-305. (pdf)
Cibois A, Kalyakin MV, Lian-Xian H, and Pasquet E (2002), Molecular phylogenetics of babblers (Timaliidae): revaluation of the genera Yuhina and Stachyris, J. Avian Biol. 33, 380-390. (free reading)
Clayton DA (1991), Replication and transcription of vertebrate mitochondrial DNA, Annu. Rev. Cell Biol. 7, 453-478. (link)
Clayton DA (1992), Transcription and replication of animal mitochondrial DNAs, Int. Rev. Cyt. 141, 217-232. (abstract)
Cobb LJ, Lee C, Xiao J, Yen K, Wong RG, Nakamura HK, Mehta HH, Gao Q, Ashur C, Huffman DM, Wan J, Muzumdar R, Barzilai N, and Cohen P (2016), Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers, AGING 8, 796-809. (pdf)
Colihueque N, Gants A, and Parraguez M (2021), Revealing the diversity of Chilean birds through the COI barcode approach, ZooKeys 1016, 143-161. (free pdf)
Cooper A, Lalueza-Fox C, Anderson S, Rambaut A, Austin J, and Ward R (2001), Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution, Nature 409, 704-707. (abstract)
Crochet PA, and Desmarais E (2000), Slow rate of evolution of the mitochondrial control region of gulls (Aves: Laridae), Mol. Biol. Evol. 17, 1797-806. (free pdf)
Cucini C, Leo C, Iannoti N, Boschi S, Brunetti C, Pons J, Fanciulli PP, Frati F, Carapelli A, and Nardi F (2021), EZmito: a simple and fast tool for multiple mitogenome analyses, Mitochondrial DNA B 6, 1101-9. (pdf)
Cui B, Ma F, Wang X, Sun Y, Yu L, Li-Ling J, and Li Q (2006), Phylogenetic analyses of Fringillidae (Aves: Passeriformes) using mitochondrial tRNA gene sequences and secondary structure, J. Biol. Syst. 14, 413-424.(abstract)
Delport W, Ferguson J, and Bloomer P (2002), Characterization and evolution of the mitochondrial DNA control region in hornbills (Bucerotiformes), J. Mol. Evol. 54, 794-806. (abstract)
Del-Rio G, Rego MA, Whitney BM, Schunck F, Silveira LF, Faircloth BC, and Brumfield RT (2021), Displaced clines in an avian hybrid zone (Thamnophilidae: Rhegmatorhina) within an Amazonian Interfluve, Evolution (abstract)
De Panis D, Lambertucci SA, Wiemeyer G, Dopazo H, Almeida FC, Mazzoni CJ, Gut M, Gut I, and Padró J (2021), Mitogenomic analysis of extant condor species provides insight into the molecular evolution of vultures, Sci. Rep. 11, e:17109. (pdf)
Desjardins P, and Morais R (1990), Sequence and gene organization of the chicken mitochondrial genome: a novel gene order in higher vertebrates, J. Mol. Biol. 212, 599-634. (abstract)
Desjardins P, and Morais R (1991), Nucleotide sequence and evolution of coding and noncommittal regions of a quail mitochondrial genome, J. Mol. Evol. 32, 153-161. (abstract)
Dey P, Sharma SK, Sarkar I, Ray SD, Pramod P, Kochiganti VHS, Quadros G, Rathore SS, Singh V, and Singh RP (2021), Complete mitogenome of endemic plum-headed parakeet Psittacula cyanocephala – characterization and phylogenetic analysis, PLOS ONE 16, e:0241098. (free pdf)
Dey P, Sarkar I, Ray SD, Pramod P, Natarajan J, and Singh RP (2022), Genome survey sequencing, microsatellite motif identification and complete mitogenome of Turnix suscitator – novel implications for Charadriiformes phylogeny, Helyon (pdf)
Dias C, de Araújo Lima K, Araripe J, Aleixo A, Vallinoto M, Sampaio I, Schneider H, and Sena do Rêgo P (2018), Mitochondrial introgression obscures phylogenetic relationships among manakins of the genus Lepidothrix (Aves: Pipridae), Mol. Phylogenet. Evol. 126, 314-320. (abstract)
Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, and Bernt M (2019), Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes, Nucleic Acids Res. 47, 10543-52. (free pdf)
Donne-Goussé C, Laudet V, and Hänni C (2002), A molecular phylogeny of anseriformes based on mitochondrial DNA analysis, Mol. Phylogenet. Evol. 23, 339-356. (abstract)
Du C, Zhang L, Lu T, MA J, Zeng C, Yue B, and Zhang AX (2017), Mitochondrial genomes of blister beetles (Coleoptera, Meloidae) and two large intergenic spacers in Hycleus genera, BMC Genomics 18, e:698. (pdf)
Du C, Liu Y, and Fu Z (2020), The complete mitochondrial genome of the Eurasian wryneck Jynx torquilla (Aves: Piciformes: Picidae) and its phylogenetic inference, Zootaxa 4810, 351-360. (abstract)
Eberhard JR, and Wright TF (2016), Rearrangement and evolution of mitochondrial genomes in parrots, Mol. Phylogenet. Evol. 94, 34-46. (abstract)
Eberhard JR, Wright TF, and Bermingham E (2001), Duplication and concerted evolution of the mitochondrial control region in the parrot genus Amazona, Mol. Biol. Evol. 18, 1330-1342. (free pdf)
Edwards SV, Jennings WB, and Shedlock AM (2005), Phylogenetics of modern birds in the era of genomics, Proc. R. Soc. B 272, 979-992. (pdf)
Falkenberg M, and Gustafsson CM (2020), Mammalian mitochondrial DNA replication and mechanisms of deletion formation, Crit. Rev. Biochem. Mol. Biol. 55, 509-524. (pdf)
Feinstein J (2006), The mitochondrial genome of Cygnus columbianus, the Whistling Swan, DNA Sequence 17, 99-106. (abstract)
Formenti G, Rhie A, Balacco J, Haase B, Mountcastle J, Fedrigo O, Brown S, Capodiferro MR, Al-Ajli FO, Ambrosini R, Houde P, Koren S, Oliver K, Smith M, Skelton J, Betteridge E, Dolucan J, Corton C, Bista I, Torrance J, Tracey A, Wood J, Uliano-Silva M, Howe K, McCarthy S, Winkler S, Kwak W, Korlach J, Fungtammasan A, Fordham D, Costa V, Mayes S, Chiara M, Horner DS, Myers E, Durbin R, Achilli A, Braun EL, Phillippy AM, and Jarvis ED (2021), Complete vertebrate mitogenomes reveal widespread repeats and gene duplications, Genome Biol. 22, e:120. (pdf)
Funk DJ, and Omland KE (2003), Species-level paraphyly and polyphyly: frequency, causes and consequences with insights from animal mitochondrial DNA, Annu. Rev. Ecol. Evol. Syst. 22, 397-422. (abstract)
Galtier N, Nabholz B, Glémin S, and Hurst GDD (2009), Mitochondrial DNA as a marker of molecular diversity: a reappraisal, Mol. Ecol. 18, 4541-50. (free pdf)
Gao J, Wang G, Zhou C, Price M, Ma J, Sun X, Chen B, Zhang X, and Yue B (2021), Complete mitochondrial genome of Fulvetta cinereiceps (Sylviidae: Passeriformes) and consideration of its phylogeny within babblers, Pakistan J. Zool. 53, 2091-2104. (pdf)
Gibb GC, Kardailsky O, Kimball RT, Braun EL, and Penny D (2007), Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations, Mol. Biol. Evol. 24, 269-280. (free pdf)
Gibb GC, Kennedy M, and Penny D (2013), Beyond phylogeny: pelecaniform and ciconiiform birds, and long-term niche stability, Mol. Phylogenet. Evol. 68, 229-238. (abstract)
Gibb GC, England R, Hartwig G, McLenachan PA, Smith BLT, McComish BJ, Cooper A, and Penny D (2015), New Zealand passerines help clarify the diversification of major songbird lineages during the Oligocene, Genome Biol. Evol. 7, 2983-95. (free pdf)
Gilbert PS, Wu J, Simon MW, Sinsheimer JS, and Alfaro ME (2018), Filtering nucleotide sites by phylogenetic signal to noise ratio increases confidence in the Neoaves phylogeny generated from ultraconserved elements, Mol. Phylogenet. Evol. 126, 116-128. (pdf)
Gissi C, Ianneli F, and Pesole G and (2008), Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species, Heredity 101, 301-320. (pdf)
Gong J, Zhao R, Huang Q, Sun X, Huang L, and Jing M (2017), Two mitogenomes of Gruiformes (Amaurornis akool/ A. phoenicurus) and the phylogenetic placement of Rallidae, Genes Genom. 39, 987-995. (abstract)
Gonzalez J, Duttmann H, and Wink M (2009), Phylogenetic relationships based on two mitochondrial genes and hybridization patterns in Anatidae, J. Zool. 279: 310-318. (abstract)
Gordon EL, Kimball RT, and Braun EL (2021), Protein structure, models of sequence evolution, and data type effects in phylogenetic analyses of mitochondrial data: a case study in birds, Diversity 13, e:555. (pdf)
Griffiths CS (1997), Correlation of functional domains and rates of nucleotide substitution in Cytochrome b, Mol. Phylogenet. Evol. 7, 352-365. (abstract)
Groth JG (1998), Molecular phylogenetics of finches and sparrows: consequences of character state removal in cytochrome b sequences, Mol. Phylogenet. Evol. 10, 377-390. (abstract)
Guan X, Silva P, Gyenai KB, Xu J, Geng T, Tu Z, Samuels DC and Smith EJ (2009), The mitochondrial genome sequence and molecular phylogeny of the turkey, Meleagris gallopavo, Animal Genet. 40, 134-141. (pdf)
Guo ZL, Zhang Y, Yang H, Wang TS, Wang WW, Zeng SS, Guo Y, Ye L, Du A, Wang ZW, Zeng SM, Tuan J, and Wang L (2022), Sequencing and structural characteristic analysis of mitochondrial genome in Zhijin White Goose (Anser cygnoides), Res. Square (pdf)
Gutell RR, Lee JC, & Cannone JJ (2002), The accuracy of ribosomal RNA comparative structure models, Curr. Opin. Struct. Biol. 12, 301-310. (abstract)
Haddrath O, and Baker AJ (2001), Complete mitochondrial DNA genome of extinct birds: ratite phylogenetics and the vicariance biogeography hypothesis, Proc. R. Soc. Lond. B 268, 239-245. (pdf)
Härlid A, Janke A, and Arnason U (1997), The mtDNA sequence of the ostrich and the divergence between paleognathous and neognathous birds, Mol. Biol. Evol. 14, 754-761. (free pdf)
Härlid A, Janke A, and Arnason U (1998), The complete mitochondrial genome of Rhea americana and early avian divergences, J. Mol. Evol. 46, 669-679. (abstract)
Halley YA, Oldeschulte DL, Bhattarai EK, Hill J, Metz RP, Johnson CD, Presley SM, Ruzicka RE, Rollins D, Peterson MJ, Murphy WJ, and Seabury CM (2015), Northern Bobwhite (Colinus virginianus) mitochondrial population genomics reveals structure, divergence, and evidence for heteroplasmy, PLoS ONE 10, e:0144913. (pdf)
Hanna ZR, Henderson JB, Sellas AB, Fuchs J, Bowie RCK, and Dumbacher JP (2017), Complete mitochondrial genome sequences of the northern spotted owl (Strix occidentalis caurina) and the barred owl (Strix varia; Aves: Strigiformes: Strigidae) confirm the presence of a duplicated control region, PeerJ 5, e:3901. (pdf)
Hansen CCR, Baleka S, Gudjónsdottir SM, Rasmussen JA, Ballesteros JAC, Hallgrimsson GT, Stefansson RA, von Schmalensee M, Skarphédinsson KH, Labansen AL, Leivits M, Skelmose, Sonne C, Dietz R, Boertmann D, Eulaers I, Martín MD, and Pálsson S (2022), Distinctive mitogenomic lineages within populations of White-tailed Eagles, Ornithology 139, (abstract)
Haring E, Riesing MJ, Pinsker W, and Gamauf A (1999), Evolution of a pseudo-control region in the mitochondrial genome of Palearctic buzzards (genus Buteo), J. Zool. Syst. Evol. Res. 37, 185-194. (abstract)
Haring E, Kruckenhauser L, Gamauf A, Riesling MJ, and Pinsker W (2001), The complete sequence of the mitochondrial genome of Buteo buteo (Aves, Accipitridae) indicates an early split in the phylogeny of raptors, Mol. Biol. Evol. 18, 1892-1904. (free pdf)
Haring E, Gamauf A, and Kryukov A (2007), Phylogeographic patterns in widespread corvid birds, Mol. Phylogenet. Evol. 45, 840-862. (abstract)
Harrison GL, McLenachan PA, Phillips MJ, Slack KE, Cooper A, and Penny D (2004), Four new avian mitochondrial genomes help get to basic evolutionary questions in the Late Cretaceous, Mol. Biol. Evol. 21, 974-983. (free pdf)
Havird JC, and Santos SR (2014), Performance of single and concatenated sets of mitochondrial genes at inferring metazoan relationships relative to full mitogenome data, PLoS ONE 9, e:84080. (pdf)
He K, Ren T, Zhu S, and Zhao A (2015), The complete mitochondrial genome of Fulica atra (Avian, Gruiformes, Rallidae), Mitochondrial DNA
He L, Dai B, Zeng B, Zhang X, Chen B, Yue B, and Li J (2009), The complete mitochondrial genome of the Sichuan Hill Partridge (Arborophila rufipectus) and a phylogenetic analysis with related species, Gene 435, 23-28. (abstract)
He XL, Ding CQ, and Han JL (2013), Lack of structural variation but extensive length polymorphisms and heteroplasmic length variations in the mitochondrial DNA control region of highly inbred Crested Ibis, Nipponia nippon, PLOS ONE 8, e:66324. (pdf)
Hebert PDN, Ratnasingham S, and deWaard JR (2003), Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species, Proc. R. Soc. Lond. B (Suppl.) 270, S96-99. (pdf)
Hebert PDN, Stoeckle MY, Zemlak TS, and Francis CM (2004), Identification of birds through DNA barcodes, PLoS Biol. 2, 1657-63. (pdf)
Hosner PA, Zhao M, Kimball RT, Braun EL, and Burleigh JG (2022), Reconciling GenBank names with standardized avian taxonomies to improve linkage between phylogeny and phenotype, Ornithology 139, e:ukac045. (abstract)
Houde P, Cooper A, Leslie E, Strand AE, and Montaño GA (1997), Phylogeny and evolution of 12S rDNA in Gruiformes (Aves), Avian Mol. Evol. Syst. 1997, 121-158. (abstract)
Hu C, Zhang C, Sun L, Zhang y, Xie W, Zhang B, and Chang Q (2017), The mitochondrial genome of pin-tailed snipe Gallinago stenura, and its implications for the phylogeny of Charadriiformes, PLOS ONE 12, e:0175244. (pdf)
Hu C, Xu X, Yao W, Liu W, Tai D, Chen W, and Chang Q (2021), Complete mitochondrial genome of the Eurasian Oystercatcher Haematopus ostralegus and comparative genomic analyses of Charadriiformes, Pakistan J. Zool. 53, 2407-15. (free pdf)
Huang Z, Liu N, Xiao Y, Cheng Y, Mei W, and Wen L (2009), Phylogenetic relationships of four endemic genera of the Phasianidae in China based on mitochondrial DNA control region genes, Mol. Phylogenet. Evol. 53, 378-383. (abstract)
Huang Z, Liu N, Liang W, Zhang Y, Liao X, Ruan L, and Yang Z (2010), Phylogeography of Chinese bamboo partridge, Bambusicola thoracica thoracica (Aves: Galliformes) in south China: inference from mitochondrial DNA control-region sequences, Mol. Phylogenet. Evol. 56, 273-280. (abstract)
Huang Z, and Ke D (2016a), Structure and evolution of the Phasianidae mitochondrial control region, Mitochondrial DNA A 27, 350-354. (abstract)
Huang Z, and KE D (2016b), Structure and variation of the Anseriformes mitochondrial DNA control region, Mitochondrial DNA A 27, 2036-39. (abstract)
Huang Z, Tu F, and Murphy RW (2016), Analysis of the complete mitogenome of Oriental turtle dove (Streptopelia orientalis) and implications for species divergence, Biochem. Syst. Ecol. 65, 209-213. (abstract)
Huang Z, and KE D (2017), Organization and variation of the Tetraonidae (Aves: Galliformes) mitochondrial DNA control region, Mitochondrial DNA B 2, 568-570. (pdf)
Huang Z, Li M, Ruan R, and Qin J (2017a), Organization and variation of the mitochondrial DNA control region in Ardeidae (Aves: Ciconiiformes) and their phylogenetic relationship, Pakistan J. Zool. 49, 1917-1920. (pdf, incl. Suppl. Mat.)
Huang Z, Shen Y, and Ma Y (2017b), Structure and variation of the Fringillidae (Aves: Passeriformes) mitochondrial DNA control region and their phylogenetic relationship, Mitochondrial DNA A 28, 867-871. (abstract)
Huang Z, Tu F, and Tang S (2018), Two new mitogenomes of Pellorneidae (Aves: Passeriformes) and a phylogeny of the superfamily Sylvioidea, Austr. J. Zool. 66, 167-173. (abstract)
Hussain T, Babar ME, Musthafa M, Wajid A, Sukhera HA, Chaudhry JI, Khan MR, and Marikar FMMT (2022), Genetic polymorphism in the mitochondrial D-loop of Oriental White-backed Vultures, Ornis hungarica 30, 135-145. (pdf)
Jemt E, Persson Ö, Shi Y, Mehmedovic M, Uhler JP, López MD, Freyer C, Gustafsson CM, Samuelsson T, and Falkenberg M (2015), Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element, Nucleic Acid Res. 43, 9262-75. (pdf)
Jia Y, Qiu G, Cao C, Wang X, Jiang L, Zhang T, Geng Z, and Jin S (2023), Mitochondrial genome and phylogenetic analysis of Chaohu duck, Gene 851, e:147018. (abstract)
Jiang F, Miao Y, Liang W, Ye H, Liu H, and Liu B (2010), The complete mitochondrial genomes of the whistling duck (Dendrocygna javanica) and black swan (Cygnus atratus): dating evolutionary divergence in Gallianserae, Mol. Biol. Rep. 37, 3001-15. (abstract)
Jiang L, Wang G, Peng R, Peng Q, & Zou F (2014), Phylogenetic and molecular dating analysis of Taiwan Blue Pheasant (Lophura swinhoii), Gene 539, 21-29. (abstract)
Jiang L, Chen J, Wang P, Ren Q, Yuan J, Qian C, Hua X, Guo Z, Zhang L, Yang J, Wang Y, Zhang Q, Ding H, Bi D, Zhang Z, Wang Q, Chen D, and Kan X (2015), The mitochondrial genomes of Aquila fasciata and Buteo lagopus (Aves, Accipitriformes): sequence, structure and phylogenetic analyses, PLoS ONE 10, e:0136297. (pdf)
Jiang L, Peng L, Tang M, You Z, Zhang M, West A, Ruan Q, Chen W, and Merilä J (2019), Complete mitochondrial genome sequence of the Himalayan Griffon, Gyps himalayensis (Accipitriformes: Accipitridae): sequence, structure, and phylogenetic analyses, Ecol. Evol. 9, 8813-28. (pdf)
Jin S, Xia J, Jia F, Jiang L, Wang X, Liu X, Liu X, and Geng Z (2022), Complete mitochondrial genome, genetic diversity and phylogenetic diversity of Pingpu Yellow Chicken (Gallus gallus), Animals 12, e:3037. (free pdf)
Jin X, Cheng Z, Wang B, Yau T, Chen Z, Barker SC, Chen D, Bu W, Sun D, and Gao S (2020), Precise annotation of human, chimpanzee, rhesus macaque and mouse mitochondrial genomes leads to insight into mitochondrial transcription in mammals, RNA Biol. 17, 395-402. (pdf)
Jing M, Yang H, Li K, and Huang L (2020), Characterization of three new mitochondrial genomes of Coraciiformes (Megaceryle lugubris, Alcedo atthis, Halcyon smyrnensis) and insights into their phylogenetics, Genet. Mol. Biol. 43, e:20190392. (pdf)
Johnsen A, Kearns AM, Omland KE, and Anmarkrud JA (2017), Sequencing of the complete mitochondrial genome of the common rave Corvus corax (Aves: Corvidae) confirms mitogenome-wide deep lineages and a paraphyletic relationship with the Chihuahuan raven C. cryptoleucus, PLoS ONE 12, e:0187316. (pdf)
Jühling F, Pütz J, Bernt M, Donath A, Middendorf M, Florentz C, and Stadler PF (2012), Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements, Nucl. Acid Res. 40, 2833-45. (pdf)
Kan XZ, Li XF, Lei ZP, Wang M, Chen L, Gao H, and Yang ZY (2010a), Complete mitochondrial genome of Cabot‘s tragopan, Tragopan caboti (Galliformes: Phasianidae), Genet. Mol. Res. 9, 1204-1216. (pdf)
Kan XZ, Li XF, Zhang LQ, Chen L, Qian CJ, Zhang XW, and Wang L (2010b), Characterization of the complete mitochondrial genome of the Rock pigeon, Columba livia (Columbiformes, Columbidae), Genet. Mol. Res. 9, 1234-49. (pdf)
Kan X, Yuan J, Zhang L, Li X, Yu L, Chen L, Guo Z, and Yang J (2013), Complete mitochondrial genome of the Tristram’s Bunting, Emberiza tristrami (Aves: Passeriformes): the first representative of the family Emberizidae with six boxes in the central conserved domain II of control region, Mitochondrial DNA 24, 648-650. (abstract)
Kang H, Li B, Ma X, and a Xu at (2018), Evolutionary progression of mitochondrial gene rearrangements and phylogenetic relationships in Strigidae (Strigiformes), Gene 674, 8-14. (abstract)
Ke Y, Huang Y, and Lei FM (2008), [Sequencing and analysis of the complete mitochondrial genome of Podoces hendersoni (Aves, Corvidae)], Yi Chuan. 32, 951-960. (Chinese) (abstract)
Kerr KCR, Stoeckle MY, Dove CJ, Weigt LA, Francis CM, and Hebert PDN (2007), Comprehensive DNA barcode coverage of North American birds, Mol. Ecol. Notes 7, 535-543. (pdf)
Kim JI, Do TD, Choi Y, Yeo Y, and Kim CB (2021), Characterization and comparative analysis of complete mitogenomes of three Cacatua parrots (Psittaciformes: Cacatuidae), Genes 12, e:209. (pdf)
Kim JI, Do TD, Yeo Y, and Kum CB ((2022), Comparative analysis of complete mitochondrial genomes of three Trichoglossus species (Psittaciformes: Psittacidae), Mol. Biol. Rep. (abstract)
Kumazawa Y, and Nishida M (1993), Sequence evolution of mitochondrial tRNA genes and deep-branch animal phylogenetics, J. Mol. Evol. 37, 380-398. (abstract)
Kundu S, Alam I, Maheswaran G, Tyagi K, and Kumar (2021), Complete mitochondrial genome of Great Frigatebird (Fregata minor): phylogenetic position and gene rearrangement, Biochem. Genet. 60, 1177-1188. (abstract)
Kuro-o M, Yonekawa H, Saito S, Eda M, Higuchi H, Koike H, and Hasegawa H (2010), Unexpectedly high genetic diversity of mtDNA control region through severe bottleneck in vulnerable albatross Phoebastria albatrus, Conserv. Genet. 11, 127-137. (abstract)
L‘Abbé D, Duhaime JF, Lang BF, and Morais R (1991), The transcription of DNA in chicken mitochondria initiates from one major bidirectional promoter, J. Biol. Chem. 266, 10844-50. (free pdf)
Labuschagne C, Kotzé A, Grobler JP, and Dalton DL (2014), The complete sequence of the mitochondrial genome of the African Penguin (Spheniscus demersus), Gene 534, 113-118. (abstract)
Li JY, Li WX, Wang AT, and Zhang Y (2021), MitoFlex: an efficient, high-performance toolkit for animal mitogenome assembly, annotation and visualization, Bioinformatics 37, 3001-3. (abstract)
Li X, Huang Y, and Lei F (2015), Comparative mitochondrial genomics and phylogenetic relationships of Crossoptilon species (Phasianidae, Galliformes), BMC Genomics 16, e:42. (pdf)
Liu D, Zhou Y, Xie C, and Hou S (2021), Mitochondrial genome of the critically endangered Baer’s Pochard, Aythya baeri, and its phylogenetic relationship with other Anatidae species, Sci. Rep. 11, e:24302. (pdf)
Liu F, Ma L, Yang C, Tu F, Xu Y, Ran J, Yue B, and Zhang X (2014), Taxonomic status of Tetraophasis obscurus and Tetraophasis szechenyii (Aves: Galliformes: Phasianidae) based on the complete mitochondrial genome, Zool. Sci. 31, 160-167. (abstract)
Liu G, Zhou LZ, and Gu CM (2012), Complete sequence and gene organization of the mitochondrial genome of scaly-sided merganser (Mergus squamatus) and phylogeny of some Anatidae species, Mol. Biol. Rep. 39, 2139-45. (abstract)
Liu G, Zhang Z, Luo L, and Xu W (2013), The complete mitochondrial genome of bean goose (Anser fabalis) and implications for Anseriformes taxonomy, PLoS ONE 8, e:63334. (pdf)
Liu G, Zhou L, Li B, and Zhang L (2014), The complete mitochondrial genome of Aix galericulata and Tadorna ferruginea: bearings on their phylogenetic position in the Anseriformes, PLoS ONE 9, e:109701. (pdf)
Liu G, Li C, Du Y, and Liu X (2017), The complete mitochondrial genome of Japanese sparrowhawk (Accipiter gularis) and the phylogenetic relationships among some predatory birds, Biochem. Syst. Ecol. 70, 116-125. (abstract)
Liu W, Hu C, Xie W, Chen P, Zhang Y, Yao R, , (2018), The mitochondrial genome of red-necked phalarope Phalaropus lobatus (Charadriiformes: Scolopacidae) and phylogeny analysis among Scolopacidae, Genes Genomics 40, 455-462. (abstract)
Livezey BC (1986), A phylogenetic analysis of recent anseriform genera using morphological characters, Auk 103, 737-754. (free pdf)
Lubbe P, Rawlence NJ, Kardailsky O, Robertson BC, Day R, Knapp M, and Dussex N (2022), Mitogenomes resolve the phylogeography and divergence times within the endemic New Zealand (Callaeidae: Passerida), Zool. J. Linn. Soc. (free pdf)
Lucchini V, and Randi E (1999), Molecular evolution of the mtDNA control-region in galliform birds, Proceed. 22th Intern. Ornithol. Congr., Durban, pp. 732-739. (pdf)
Lucchini V, Höglund J, Klaus S, Swenson J, and Randi E (2001), Historical biogeography and a mitochondrial DNA phylogeny of grouse and phylogenetic ptarmigan, Mol. Phylogenet. Evol. 20, 149-162. (abstract)
Ma YG, Huang Y, and Lei FM (2014), Sequencing and phylogenetic analysis of the Pyrgilauda ruficollis (Aves, Passeridae) complete mitochondrial genome, Zool. Res. 35, 81-91. (pdf)
Mackiewicz P, Urantówka AD, Kroczak A, and Mackiewicz D (2019), Resolving phylogenetic relationships within Passeriformes based on mitochondrial genes and inferring the evolution of their mitogenomes in terms of duplications, Genome Biol. Evol. 11, 2824-49. (free pdf)
Marshall HD, and Baker AJ (1997), Structural conservation and variation in the mitochondrial control region of fringilline finches (Fringilla spp.) and the Greenfinch (Carduelis chloris), Mol. Biol. Evol. 14, 173-184. (pdf)
McCracken KG, and Sorenson MD (2005), Is homoplasy or lineage sorting the source of incongruent mtDNA and nuclear gene trees in the stiff-tailed ducks (Nomonyx-Oxyura)?, Syst. Biol. 54, 35-55. (free pdf)
Meiklejohn KA, Danielson MJ, Faircloth BC, Glenn TC, Braun EL, and Kimball RT (2014), Incongruence among different mitochondrial regions: a case study using complete mitogenomes, Mol. Phylogenet. Evol. 78, 314-323. (abstract)
Meyer CP, and Paulay G (2005), DNA barcoding: error rates based on comprehensive sampling, PLoS Biol. 3, e:422. (pdf)
Miller HC, Lambert DM (2006), A molecular phylogeny of New Zealand’s Petroica (Aves: Petroicidae) species based on mitochondrial DNA sequences, Mol. Phylogenet. Evol. 40, 844-855. (abstract)
Mindell DP, Sorenson MD, Huddleston CJ, Miranda HC, Knight A, Sawchuk SJ, and Yuri T (1997), Phylogenetic relationships among and within select avian orders based on mitochondrial DNA, In: „Avian Molecular Evolution and Systematics” (Mindell DP, ed.), chapter 8. (abstract)
Mindell DP, Sorenson MD, and Dimcheff DE (1998a), Multiple independent origins of mitochondrial gene order in birds, Proc. Natl. Acad. Sci. USA 95, 10693-97. (pdf)
Mindell DP, Sorenson MD, and Dimcheff DE (1998b), An extra nucleotide is not translated in mitochondrial ND3 of some birds and turtles, Mol. Biol. Evol. 15, 1568-71. (free pdf)
Mindell DP, Sorenson MD, Dimcheff DE, Hasegawa M, Ast JC, and Yuri (1999), Interordinal relationships of birds and other reptiles based on whole mitochondrial genomes, Syst. Biol. 48, 138-152. (free pdf)
Mitchell KJ, Wood JR, Scofield RP, Llamas B, and Cooper A (2013), Ancient mitochondrial genome reveals unsuspected taxonomic affinity of the extinct Chatham duck (Pachyanas chathamica) and resolves divergence times for New Zealand and sub-Antarctic brown teals, Mol. Phylogenet. Evol. 70, 420-428. (abstract)
Mitchell KJ, Wood JR, Llamas B, McLenachan PA, Kardailsky O, Scofield RP, Worthy TH, and Cooper A (2016), Ancient mitochondrial genomes clarify the evolutionary history of New Zealand’s enigmatic acanthisittid wrens, Mol. Phylogenet. Evol. 102, 295-304. (abstract)
Montaña-Lozano P, Moreno-Carmona M, Ochoa-Capera M, Medina NS, Boore JL, and Prada CF (2022), Comparative genomic analysis of vertebrate mitochondria reveals a differential of rearrangement rate between taxonomic class, Sci. Rep. 12, e:5479. (pdf)
Morgan-Richards M, Bulgarella M, Sivyer L, Dowle EJ, Hale M, McKean NE, and Trewick SA (2017), Explaining large mitochondrial sequence differences within a population sample, R. Soc. open sci. 4, e:170730. (pdf)
Morinha F, Clemente C, Cabral JA, Lewicka MM, Travassos P, Carvalho D, Dávila JA, Santos M, Blanco G, and Bastos E (2016), Next-generation sequencing and comparative analysis of Pyrrhoxorax pyrrhoxorax and Pyrrhoxorax graculus (Passeriformes:Corvidae) mitochondrial genomes, Mitochondrial DNA 27, 2278-81. (abstract)
Morris-Pocock JA, Taylor SA, Birt TP, and Friesen VL (2010), Concerted evolution of duplicated mitochondrial control regions in three related seabird species, BMC Evol. Biol. 10, e:14. (pdf)
Moum T, and Bakke I (2001), Mitochondrial control region structure and single site heteroplasmy in the razorbill (Alca torda; Aves), Curr. Genet. 39, 198-203. (abstract)
Mu CY, Huang ZY, Chen Y, Wang B, Su YH, Li Y, Sun ZM, Xu Q, Zhao WM, and Chen GH (2014), Completely sequencing and gene organization of the Anser cygnoides mitochondrial genome, J. Agric. Biotechnol. 22, 1482-93. [in Chinese] (free pdf)
Nabholz B, Jarvis E, and Ellegren H (2010), Obtaining mtDNA genomes from next-generation transcriptome sequencing: a case study on the basal Passerida (Aves: Passeriformes) phylogeny, Mol. Phylogenet. Evol. 57, 466-470. (abstract)
Nicholls TJ, and Minczuk M (2014), In D-loop: 40 years of mitochondrial 7S DNA, Exp. Geront. 56, 175-181. (abstract)
Nishibori M, Hayashi T, Tsudzuki M, Yamamoto Y, and Yasue H (2001), Complete sequence of the Japanese quail (Coturnix japonica) mitochondrial genome and its genetic relationship with related species, Animal Genet. 32, 380-385. (abstract)
Nishibori M, Tsudzuki M, Hayashi T, Yamamoto Y, and Yasue H (2002), Complete nucleotide sequence of the Coturnix chinensis (Blue-breasted Quail) mitochondrial genome and phylogenetic analysis with related species, J. Hered. 93, 439-444. (free pdf)
Nishibori M, Hayashi T, and Yasue H (2004), Complete nucleotide sequence of Numida meleagris (Helmeted guineafowl) mitochondrial genome, J. Poultry Sci. 41, 259-268. (pdf)
Nishibori M, Shimogiri T, Hayashi T, and Yasue H (2005), Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius, Anim. Genet. 36, 367-375. (abstract)
Omote K, Nishida C, Dick MH, and Masuda R (2013), Limited phylogenetic distribution of a long tandem-repeat cluster in the mitochondrial control region in Bubo (Aves, Strigidae) and cluster variation in Blakiston’s fish owl (Bubo blakistoni), Mol. Phylogenet. Evol. 66, 889-897. (abstract)
Pacheco MA, Battistuzzi FU, Lentino M, Aguilar RF, Kumar S, and Escalante AA (2011), Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders, Mol. Biol. Evol. 28, 1927-42. (free pdf)
Päckert M (2022), Free access of published DNA sequences facilitates regular control of (meta-) data quality – an example from shorebird mitogenomes (Aves, Charadriiformes: Charadrius), Ibis 164, 336-342. (pdf)
Patton J, and Avise J (1986), Evolutionary genetics of birds IV: rates of protein divergence in waterfowl (Anatidae), Genetica 68, 129-143. (abstract)
Peng C, Li J, Lu H, Liu W, and Zhang J (2022), Characterization of the complete mitochondrial genome of the Sea Duck Mergus serrator and comparison with other Anseriformes species, Res. Square (pdf)
Pereira F, Soares P, Carneiro J, Pereira L, Richards MB, Samuels DC, and Amorim A (2008), Evidence for variable selective pressures at a large secondary structure of the hunan mitochondrial DNA control region, Mol. Biol. Evol. 25, 2759-70. (free pdf)
Pereira SL, Grau ET, and Wayntal A and (2004), Molecular architecture and rates of DNA substitutions of the mitochondrial control region of cracid birds, Genome 47, 535-545. (abstract)
Peters JL, McCracken KG, Zhuravlev YN, Lu Y, Wilson RE, Johnson KP, and Omland KE (2005), Phylogenetics of wigeons and allies (Anatidae: Anas): the importance of sampling multiple loci and multiple individuals, Mol. Phylogenet. Evol. 35, 209-224. (abstract)
Peters JL, Bolender KA, and Pearce JM (2012), Behavioural vs. molecular sources of conflict between nuclear and mitochondrial DNA: the role of male-biased dispersal in a Holarctic sea duck, Mol. Ecol. 21, 3562-75. (abstract)
Pons JM, Hassanin A, and Crochet PA (2005), Phylogenetic relationships within the Laridae (Charadriiformes: Aves) inferred from mitochondrial markers, Mol. Phylogenet. Evol. 37, 686-699. (pdf)
Portik DM, and Wiens JJ (2021), Do alignment and trimming methods matter for phylogenomic (UCE) analyses? Syst. Biol. 70, 440-462. (free pdf)
Powell AFLA, Barker FK, and Lanyon SM (2013), Empirical evaluation of partitioning schemes for phylogenetic analyses of mitogenomic data: an avian case study, Mol. Phylogenet. Evol. 66, 69-79. (abstract)
Qian C, Ren Q, Kan X, Guo Z, Yang J, Li X, Yuan J, Qian M, Zhu Q, and Zhang L (2012), Complete mitochondrial genome of the Red-billed Starling, Sturnus sericeus (Aves: Passeriformes): the first representative of the family Sturnidae with a single control region, Mitochondrial DNA 24, 129-131. (abstract)
Qian C, Wang Y, Guo Z, Yang J, and Kan X (2013), Complete mitochondrial genome of Skylark, Alauda arvensis (Aves: Passeriformes): the first representative of the family Alaudidae with two extensive heteroplasmic control regions, Mitochondrial DNA 24, 246-248 (abstract)
Que GCL, Fontanilla IKC, Widmann P, and Widmann IDL (2022), Phylogenetic placement of the Philippine Cockatoo Cacatua haematuropygia (P.L.S. Muller 1776), Sci. Diliman 34, 5-37. (pdf)
Questiau S, Eybert MC, Gaginskaya AR, Gielly L, and Taberlet P (1998), Recent divergence between two morphologically differentiated subspecies of bluethroat (Aves: Muscicapidae: Luscinia svecica) inferred from mitochondrial DNA sequence variation, Mol. Ecol. 7, 239-245. (pdf)
Quinn TW, and Wilson AC (1993), Sequence evolution in and around the mitochondrial control region in birds, J. Mol. Evol. 37, 417-425. (abstract)
Ramirez V, Savoie P, and Morais R (1993), Molecular characterization and evolution of a duck mitochondrial genome, J. Mol. Evol. 37, 296-310. (abstract)
Randi E, and Lucchini V (1998), Organization and evolution of the mitochondrial DNA control region in the avian genus Alectoris, J. Mol. Evol. 47, 449-462. (abstract)
Randi E, Lucchini V, Hennache A, Kimball RT, Braun EL, and Ligon JD (2001), Evolution of the mitochondrial DNA control region and cytochrome b genes and the inference of phylogenetic relationships in the avian genus Lophura (Galliformes), Mol. Phylogenet. Evol. 9, 187-201. (abstract)
Ratnasingham S, and Hebert PDN (2013), A DNA-based registry for all animal species: the Barcode Index Number (BIN) system, PLOS ONE 8, e:66213. (pdf)
Rawlence NJ, Scofield RP, Spencer HG, Lalas C, Easton LJ, and Alan jD (2016), Genetic and morphological evidence for two species of Leucocarbo shag (Aves, Pelecaniformes, Phalacrocoracidae) from southern South Island of New Zealand, Zool. J. Linn. Soc. 177, 676-694. (free pdf)
Ren Q, Qian C, Yuan J, Li X, Yang J, Wang P, Jiang L, Zhang Q, Wang Y, and Kan X (2014a), Complete mitochondrial genome of the Black-capped Bulbul, Pycnonotus melanicterus (Passeriformes: Pycnonotidae), Mitochondrial DNA A 27, 1378-80. (abstract)
Ren Q, Yuan J, Ren L, Zhang L, Zhang L, Jiang L, Chen D, Kan X, and Zhang B (2014b), The complete mitochondrial genome of the yellow-browed bunting, Emberiza chrysophrys (Passeriformes: Emberizidae), and phylogenetic relationships within the genus Emberiza, J. Genet. 93, 699-707. (abstract)
Ren Z, Liang Y, Su X, and Wen J (2019), Complete mitochondrial genome of Chrysolophus pictus (Galliformes: Phasianidae), a protected and endangered pheasant species of China, Conservation Genet. Resour. 11, 121-124. (abstract)
Ritchie P, and Lambert D (2000), A repeat complex in the mitochondrial control region of Adélie penguins from Antarctica, Genome 43, 613-618. (abstract)
Román-Palacios C (2023), The ‘phruta R’ package and ‘salphycon’ shiny app: increasing access, reproducability, and transparency in phylogenetic analyses, bioRxiv (pdf)
Roques S, Godoy A, Negro JJ, and Hiraldo F (2004), Organization and variation of the mitochondrial control region in two vulture species, Gypaetus barbatus and Neophron percnopterus, J. Hered. 95, 332-337. (free pdf)
Rubinoff D, and Holland BS (2005), Between two extremes: mitochondrial DNA is neither the panacea nor the nemesis of phylogenetic and taxonomic inference, Syst. Biol. 54, 952-961. (free pdf)
Ruokonen M, Kvist L, and Lumme J (2000), Close relatedness between mitochondrial DNA from seven Anser goose species, J. Evol. Biol. 13, 532-540. (pdf)
Ruokonen M, and Kvist L (2002), Structure and evolution of the avian mitochondrial control region, Mol. Phylogenet. Evol. 23, 422-432. (abstract)
Ryu SH, and Hwang UW (2012), Complete mitochondrial genome of Saunders’s gull Chroicocephalus saundersi (Charadriiformes, Laridae), Mitochondrial DNA 23, 134-136. (abstract)
Saccone C, Pesole G, and Sbisá E (1991), The main regulatory region of the mammalian mitochondrial DNA: structure-function model and evolutionary pattern, J. Mol. Evol. 33, 83-91. (abstract)
Sammler S, Bleidorn C, and Tiedemann R (2011), Full mitochondrial genome sequences of two endemic Philippine hornbill species (Aves: Bucerotidae) provide evidence for pervasive mitochondrial DNA recombination, BMC Genomics 12, e:35. (pdf)
Sammler S, Keramiker V, Havenstein K, Krause U, Curio E, and Tiedemann R (2012), Mitochondrial control region I and microsatellite analyses of endangered Philippine hornbill species (Aves; Bucerotidae) detect gene flow between island populations and genetic diversity loss, BMC Evol. Biol. 203, e:203. (pdf)
Sammler S, Keramiker V, Havenstein K, and Tiedemann R (2013), Intraspecific rearrangement of duplicated mitochondrial control regions in the Luzon Tarictic Hornbill Penelopides manillae (Aves: Bucerotidae), J. Mol. Evol. 77, 199-205. (abstract)
Sangster G, and Luksenburg JA (2021), Sharp increase of problematic mitogenomes of birds: causes, consequences and remedies, Genome Biol. Evol. 13, e:evab210. (free pdf)
Sarkar I, Dey P, Sharma SK, Ray SD, Kochiganti VHS, Singh R, Pramod P, and Singh RP (2020), Turdoides affinis mitogenome reveals the translational efficiency and importance of NADH dehydrogenase complex-I in the Leiotrichidae family, Sci. Rep. 10, e:16202. (pdf)
Saunders MA, and Edwards SV (2000), Dynamics and phylogenetic implications of mtDNA control region sequences in New World jays (Aves: Corvidae), J. Mol. Evol. 51, 97-109. (abstract)
Sbisà E, Tanzariello F, Reyes A, Pesole G, and Saccone C (1997), Mammalian mitochondrial D-loop region structural analysis: identification of new conserved sequences and their functional and evolutionary implications, Gene 205, 125-140. (abstract)
Schirtzinger EE, Tavares ES, Gonzales LA, Eberhard JR, Miyaki CY, Sanchez JJ, Hernandez A, Müeller H, Graves GR, Fleischer RC, and Wright T (2012), Multiple independent origins of mitochondrial control region duplications in the order Psittaciformes, Mol. Phylogenet. Evol. 64, 342-356. (pdf)
Seibold I, and Helbig AJ (1995), Evolutionary history of New and Old World vultures inferred from nucleotide sequences of the mitochondrial cytochrome b gene, Philos. Trans. R. Soc. Lond. B 350, 163-178. (abstract)
Seligmann H, and Labra A (2014), The relation between harpin formation by mitochondrial WANCY tRNAs and the occurrence of the light strand replication origin in Lepidosauria, Gene 542, 248-257. (abstract)
Seutin G, Lang BF, Mindell DP, and Morais R (1994), Evolution of the WANCY region in amniotic mitochondrial DNA, Mol. Biol. Evol. 11, 329-340. (free pdf)
Shen YY, Liang L, Sun YB, Yue BS, Yang XJ, Murphy RW, and Zhang YP (2010), A mitogenomic perspective on the ancient, rapid radiation in the Galliformes with an emphasis on the Phasianidae, BMC Evol. Biol. 10, e:132. (pdf)
Shokolenko IN, and Alexeyev MF (2017), Mitochondrial transcription in mammalian cells, Front. Biosci. 22, 835-853. (pdf)
Shuangye W, Yunlin Z, Zhenggang X, Tian H, Guiyan Y, and Zhiyuan H (2021), The complete mitochondrial genome comparison between Pelecanus occidentalis and Pelecanus crispus, Russ. J. Genet. 57, 1073-81. (abstract)
Singh TR, Shneor O, and Huchon D (2008), Bird mitochondrial gene order: insight from 3 warbler mitochondrial genomes, Mol. Biol. Evol. 25, 475-477. (free pdf)
Slack KE, Janke A, Penny D, and Arnason U (2003), Two new avian mitochondrial genomes (penguin and goose) and a summary of bird and reptile mitogenomic features, Gene 302, 43-52. (abstract)
Sloan DB, Havird JC, and Sharbrough J (2016), The on-again, off-again relationship between mitochondrial genomes and species boundaries, Mol. Ecol. 26, 2212-36. (pdf)
Soares AER, Novak BJ, Haile J, Heupink TH, Fjeldså J, Gilbert MTP, Poinar H, Church GM, and Shapiro B (2016), Complete mitochondrial genomes of living and extinct pigeons revise the timing of the columbiform radiation, BMC Evol. Biol. 16, e:230. (pdf)
Song X, Huang J, Yan C, Xu G, Zhang X, and Yue B (2015), The complete mitochondrial genome of Accipiter virgatus and evolutionary history of the pseudo-control regions in Falconiformes, Biochem. Syst. Ecol. 58, 75-84. (abstract)
Spicer GS, and Dunipace L (2004), Molecular phylogeny of songbirds (Passeriformes) inferred from mitochondrial 16S RNA gene sequences, Mol. Phylogenet. Evol. 30, 325-335. (abstract)
Spiridonova LN, and Surmach SG (2018), Whole mitochondrial genome of Blakiston’s fish owl Bubo (Ketupa) blakistoni suggests its redesription in the genus Ketupa, Russ. J. Genet. 54, 369-373. (abstract)
Spiridonova LN, Valchuk OP, and Red‘kin YA (2019) A new case of recombination between nuclear and mitochondrial genomes in the genus Calliope Gould, 1836 (Muscicapidae, Aves): the hypothesis of origin Calliope pectoralis Gould 1837. Russ. J. Genet. 55, 89-99. (abstract)
Spiridonova LN, and Valchuk OP (2022), Mitochondrial genome of Phylloscopus examinandus and hypothesis of its origin, Russ. J. Genet. 58, 365-368. (abstract)
Stellbrink B, Von Rintelen T, Richter K, Finstermeier K, Frahnert S, Cracraft J, and Hofreiter M (2022), Insights into the geographical origin and phylogeographical patterns of Paradisaea birds-of-paradise, Zool. J. Linn. Soc. 196, 1394-1407. (abstract)
Stoneking M (2000), Hypervariable sites in the mtDNA control region are mutational hotspots, Am. J. Hum. Genet. 67, 1029-32. (pdf)
Sun CH, Liu HY, Min X and Lu CH (2020), Mitogenome of the little owl Athene noctua and phylogenetic analysis of Strigidae, Int. J. Biol. Macromol. 151, 924-931. (abstract)
Sun G, Zhao C, Xia T, Wei Q, Yang X, Feng S, Sha W, and Zhang H (2020), Sequence and organisation of the mitochondrial genome of Japanese Grosbeak (Eophona personata), and the phylogenetic relationships of Fringillidae, ZooKeys 995, 67-80. (free pdf)
Sun X, Zhao R, Zhang T, Gong J, Jung M, and Huang L (2017), Two mitochondrial genomes in Alcedinidae (Ceryle rudis/Halcyon pileata) and the phylogenetic placement of Coraciiformes, Genetica 145, 431-440. (abstract)
Taanman JW (1999), The mitochondrial genome: structure, transcription, translation and replication, Biochim. Biophys. Acta 1410, 103-123. (free reading)
Taylor RS, Bramwell AC, Clemente-Carvalho R, Cairns NA, Bonier F, Dares K, and Lougheed SC (2021), Cytonuclear discordance in the crowned-sparrows, Zonotrichia atricapilla and Zonotrichia leucophrys, Mol. Phylogenet. Evol. 162, e:107216. (free reading)
Thomas S, Singha HS, Kamalakkannan R, Gaughan S, Bhavana K, and Nagarajan M (2021), Analysis of the mitochondrial genome of the Indian darter, Anhinga melanogaster, suggests a species status taxonomic rank, Mol. Biol. Rep. 48, 7343-50. (abstract)
Tizard J, Patel S, Waugh J, Tavares E, Bergmann T, Gill B, Norman J, Christidis S, Scofield P, Haddrath O, Baker A, Lambert D, and Millar C (2019), DNA barcoding a unique avifauna: an important tool for evolution, systematics and conservation, BMC Evol. Biol. 19, e:52. (pdf)
Uddin A, and Chakraborty S (2015), Synonymous codon usage pattern in mitochondrial CYB gene in pisces, aves, and mammals, Mitochondrial DNA A, 187-196. (abstract)
Urantówka AD, Kroczak A, Silva T, Padrón RZ, Gallardo NF, Blanch J, Blanch B, and Mackiewicz P (2018), New insight into parrot’s mitogenomes indicates that their ancestor contained a duplicated region, Mol. Biol. Evol. 35, 2989-3008. (free pdf)
Urantówka AD, Kroczak A, and Mackiewicz P (2020), New view on the organization and evolution of Palaeognathae mitogenomes poses the question on the ancestral rearrangement in Aves, BMC Genomics 21, e:874. (pdf)
Urantówka AD, Kroczak A, Strzala T, Zaniewicz G, Kurkowski M, and Mackiewicz P (2021), Mitogenomes of Accipitriformes and Cathartiformes were subjected to ancestral and recent duplications followed by gradual degradation, Genome Biol. Evol. 13, e:evab193. (pdf)
Ushida A, Murugesapillai D, Kastner M, Wang Y, Lodeiro MF, Prabhakar S, Oliver GV, Arnold JJ, Maher LJ, Williams MC, and Cameron CE (2017), Unexpected sequences and structures of mtDNA required for efficient transcription from the first heavy-strand promoter, eLife 6, e:27283. (pdf)
Väli Ü, Treinys R, Bergmanis U, Daroczi S, Demerdzhiev D, Donbrovski V, Dravecky M, Ivanovski V, Kicko J, Langgemach T, Lontkowski J, Maciorowski G, Poirazidis K, Rodziewicz M, and Meyburg BU (2022), Contrasting patterns of genetic diversity and lack of population structure in the lesser spotted eagle Clanga pomarina (Aves: Accipitriformes) across its breeding range, Biol. J. Linn. Soc. (abstract)
Valverde JR, Marco R, and Garesse R (1994), A conserved heptamer motif for ribosomal RNA transcription termination in animal mitochondria, Proc. Natl. Acad. Sci. USA 91, 5368-71. (pdf)
van den Burg MP, and Vieites DR (2022), Bird genetic databases need improved curation and error reporting to NCBI, Ibis 165, 472-481 (pdf)
Verkuil YI, Piersma T, and Baker AJ (2017), A novel mitochondrial gene order in shorebirds (Scolopacidae, Charadriiformes), Mol. Phylogenet. Evol. 57, 411-416. (abstract)
Wägele JW (1996), Identification of apomorphies and the role of groundpatterns in molecular systematics, J. Zoo. Syst. Evol. Research 34, 31-39. (pdf)
Wang E, Zhang D, Braun MS, Hotz-Wagenblatt A, Pärt T, Arlt D, Schmaljohann H, Bairlein F, Lei F, and Wink M (2020), Can mitogenomes of the Northern Wheatear (Oenanthe oenanthe) reconstruct its phylogeography and reveal the origin of migrant birds?, Sci. Rep. 10, e:9290. (pdf)
Wang N, Hosner PA, Liang B, Braun EL, and Kimball RT (2017), Historical relationships of three enigmatic phasianid genera (Aves: Galliformes) inferred using phylogenomic and mitogenomic data, Mol. Phylogenet. Evol. 109, 217-225. (abstract)
Wang X, Liu N, Zhang H, Yang XY, Huang Y, and Lei F (2015), Extreme variation in patterns of tandem repeats in mitochondrial control region of yellow-browed tits (Sylviparus modestus, Paridae), Sci. Rep. 5, e:13227. (pdf)
Wang Y, Zhan H, Zhang Y, Long Z, and Yang X (2023), Mitochondrial genome analysis and phylogeny and divergence time evaluation of the Strix aluco, Biodiv. Data J. 11, e:101942. (pdf)
Warzecha J, Fornal A, Oczkowicz M, and Bugno-Poniewierska M (2018), A molecular characteristic of the Anatidae mitochondrial control region – a review, Ann. Anim. Sci. 18, 3-15. (pdf)
Watanabe M, Nikaido M, Tsuda TT, Kobayashi T, Mindell D, Cao Y, Okada N, and Hasegawa M (2006), New candidate species most closely related to penguins, Gene 378, 65-73. (abstract)
Wenink PW, Baker AJ, Tilanus MGJ (1994), Mitochondrial control-region sequences of two shorebird species, the Turnstone and the Dunlin, and their utility in population genetic structures, Mol, Biol. Evol. 11, 22-31. (free pdf)
Xiao B, Ma F, Sun Y, and Li QW (2006), Comparative analysis of complete mitochondrial DNA control region of four species of Strigiformes, Acta Genet. Sin. 33, 965-974. (abstract)
Xu N, Ding J, Que Z, Xu W, Ye W, and Liu H (2021), The mitochondrial genome and phylogenetic characteristics of the Thick-billed Green-Pigeon, Treron curvirostra: the first sequence for the genus, ZooKeys 1041, 167-182. (pdf)
Xu Z, Wu L, Chen J, Zhao Y, Han C, Huang T, and Yang G (2022), Insight into the characteristics of an important evolutionary model bird (Geospiza magnirostris) mitochondrial genome through comparison, Biocell 46, 1733-1746. (pdf)
Yamamoto Y, Murata K, Matsuda H, Hosoda T, Tamura K, and Furuyama JI (2000), Determination of the complete nucleotide sequence and haplotypes in the D-loop region of the mitochondrial genome in the oriental white stork, Ciconia boyciana, Genes Genet. Syst. 75, 25-32. (pdf)
Yan C, Mou B, Meng Y, Tu F, Fan Z, Price M, Yue B, and Zhang X (2017), A novel mitochondrial genome of Arborophila and new insight into Arborophila evolutionary history, PLoS ONE 12, e:0181649. (pdf)
Yang C, Lei FM, and Huang Y (2010), Sequencing and analysis of the complete mitochondrial genome of Pseudopodoces humilis (Aves, Paridae), Zool. Res. 31, 333-344. (free pdf)
Yang C, Lian T, Wang QX, Huang Y, and Xiao H (2015), Structural characteristics of the Relict Gull (Larus relictus) mitochondrial DNA control region and its comparison to other Laridae, Mitochondrial DNA 27, 2487-91. (abstract)
Yang C, Yang M, Wang Q, Lu Y, and Li X (2018), The complete mitogenome of Falco amurensis (Falconiformes, Falconidae), and a comparative analysis of genus Falco, Zool. Sci. 35, 367-372. (abstract)
Yang C, Zhao L, Wang Q, Yuan H, Li X, and Wang Y (2021), Mitogenome of Alaudala cheleensis (Passeriformes: Alaudidae) and comparative analyses of Sylvioidea mitogenomes, Zootaxa 4952, 331-353. (abstract)
Yang C, Du X, Liu Y, Yuan H, Wang Q, Hou X, Gong H, Wang Y, Huang Y, Li X, and Ye H (2022), Comparative mitogenomics of the genus Motacilla (Aves, Passeriformes) and its phylogenetic implications, ZooKeys 1109, 49-65. (free pdf)
Yang R, Wu X, Yan P, Su X, and Yang B (2010), Complete mitochondrial genome of Otis tarda (Gruiformes, Otididae) and phylogeny of Gruiformes inferred from mitochondrial DNA sequences, Mol. Biol. Rep. 37, 3057-3066. (abstract)
Yoon BK, Cho CU, and Park YC (2015), The mitochondrial genome of the Saunders’s gull Chroicocephalus saundersi (Charadriiformes: Laridae) and a higher phylogeny of shorebirds (Charadriiformes), Gene 572, 227-236. (abstract)
Yu J, Liu J, Li C, Wu W, Feng F, Wang Q, Ying X, Qi D, and Qi G (2021), Characterization of the complete mitochondrial genome of Otus lettia: exploring the mitochondrial evolution and phylogeny of owls (Strigiformes), Mitochondrial DNA B 6, 3443-51. (pdf)
Yuan QM, Luo X, Cao J, and Duan YB (2021), Complete mitochondrial genomes of three nuthatches from the genus Sitta (Aves: Passeriformes: Sittidae) and mitogenome-based phylogenetic analysis, Res. Square (pdf)
Yuan Q Sha J, and Duan Y (2022), The new mitogenome of Erpornis zantholeuca (Aves: Passeriformes): sequence, structure, and phylogenetic analysis, Cytogenet. Genome Res. 162, 250-261. (abstract)
Yuan Q, Guo Q, Cao J, Luo X, and Duan Y (2023), Description of the three complete mitochondrial genomes of Sitta (S. himalayensis, S. nagaensis, S. yunannensis) and phylogenetic relationship (Aves: Sittidae), Genes 14, e:589. (pdf)
Zhang H, Bai Y, Shi X, Wang Z, and Wu X (2018), The complete mitochondrial genomes of Tarsiger cyanurus and Phoenicurus auroreus: a phylogenetic analysis of Passeriformes, Genes Genomics 40, 151-165. (abstract)
Zhang L, Wang L, Gouda V, Wang M, Li X, and Kan X (2012), The mitochondrial genome of the Cinnamon Bittern, Ixobrychus cinnamomeus (Pelecaniformes: Ardeidae): sequence, structure and phylogenetic analysis. Mol. Biol. Rep. 39, 8315-26. (abstract)
Zhang L, Xia T, Gao X, Yang X, Sun G, Zhao C, Liu G, and Zhang H (2022), Characterization and phylogenetic analysis of the complete mitochondrial genome of Aythya marila, Res. Square (pdf)
Zhang Y, Song T, Pan T, Sun X, Sun Z, Qian L, and Zhang B (2015), Complete sequence and gene organization of the mitochondrial genome of Asio flammeus (Strigiformes, Strigidae), Mitochondrial DNA 132, 1-5. (abstract)
Zhao Q, Xu HL, and Yao YF (2018), The complete mitochondrial genome and phylogeny of the Emei Shan liocichla (Liocichla omeiensis), Conservation Genet. Resour. 11, 303-407. (abstract)
Zhao Z, Alimo Z, Zhao X, Qin H, Dayananda B, Jiang L, and Chen W (2022), Complete mitochondrial genome of Turdus merula (Aves: Passeriformes: Turdidae) and related species: genome characteristics and phylogenetic relationships, Pakistan J. Zool. (pdf)
Zhenggang X, Liang W, Sihan H, Chongxuan H, Tian H, and Yunlin Z (2021), Structural variation and phylogenetic relationship of Geospiza magnirostris based on mitochondrial control region, Biologia 76, 1367-73. (abstract)
Zhong Y, Zhou M, Ouyang B, Zeng C, Zhang M, and Yang J (2020), Complete mtDNA genome of Otus sunia (Aves, Strigidae) and the relaxation of selective constrains on Strigiformes mtDNA following evolution, Genomics 112, 3815-25. (pdf)
Zhou C, Hao Y, Ma J, Zhang W, Chen Y, Chen B, Zhang X, and Yue B (2017), The first complete mitogenome of Picumnus innominatus (Aves, Piciformes, Picidae) and phylogenetic inference within the Picidae, Biochem. Syst. Ecol. 70, 274-282. (abstract)
Zhou T, Zhou F, Yao Y, Xie M, Ni Q, Zhang M, and Xu H (2022), Comparative analysis of the complete mitochondrial genomes of related species Chrysolophus amherstiae and Chrysolophus pictus, Mitochondrial DNA B 7, 144-152. (pdf)
Zhou X, Lin Q, Fang W, and Chen X (2014), The complete mitochondrial genomes of sixteen ardeid birds revealing the evolutionary process of the gene rearrangements, BMC Genomics 15, e:573. (pdf)
Zhou XP, Lin QX, Fang WZ, and Chen XL (2014), The complete mitochondrial genomes of sixteen ardeid birds revealing the evolutionary process of the gene rearrangements, BMC Genomics 15, 1-9. (pdf)
Zimmer R, Erdtmann B, Thomas WK, and Quinn TW (1994), Phylogenetic analysis of the Coscoroba coscoroba using mitochondrial srRNA gene sequences, Mol. Phylogenet. Evol. 3, 85-91. (abstract)
Zink RM, and Barrowclough GF (2008), Mitochondrial DNA under siege in avian phylogeography, Mol. Ecol. 17, 2107-21. (pdf)
Zou Y, Jing MD, Bi XX, Zhang T, and Huang L (2015), The complete mitochondrial genome sequence of the little egret (Egretta garzeta), Genet. Mol. Biol. 38, 162-172. (pdf)

