Galliformes (landfowl)
The taxon comprises ~300 species in at least five families.
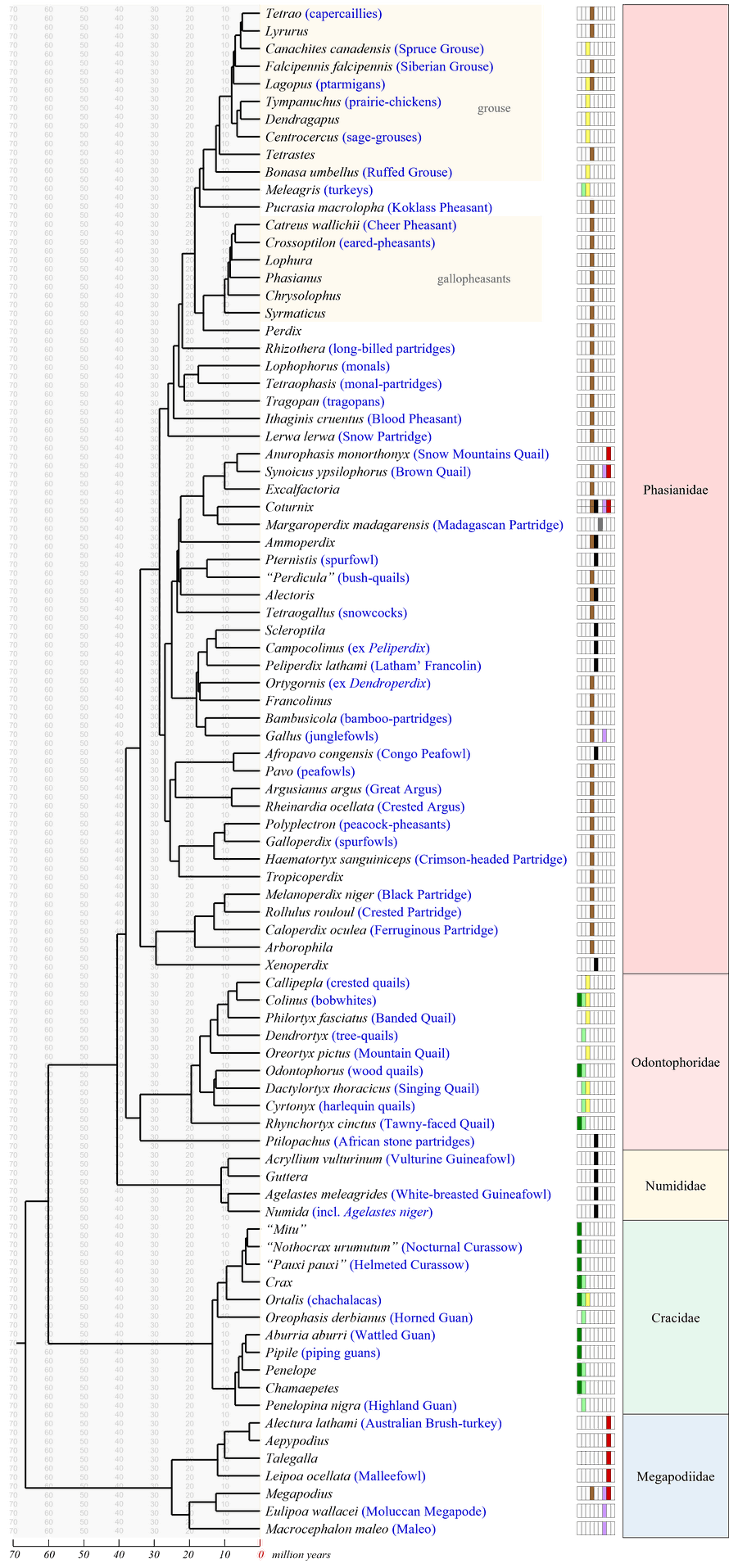
Traditional genus-level timetree of extant Galliformes including distribution data (Distribution code) according to Chen et al. (2021). Divergence times have been (slightly) adjusted to conform to the results of Stiller et al. (2024): Alectura + (Penelope + (Numida + (((Odontophorus + (Callipepla + Colinus)) + ((Coturnix + Gallus) + (Phasianus + (Meleagris + Tympanuchus)))))). [Chronoclassification would recognise two additional families, Ptilopachidae and Rollulidae.]

Traditional genus-level classification of extant Galliformes, following AviList checklist v2025. (link) The number of subspecies is given in parentheses.
References
Bai W, Zhu J, and Ren ZM (2020), Complete mitochondrial genome of Chrysolophus pictus (Galliformes: Phasianidae), a national protected wild pheasant, Mitochondrial DNA Part B 5, 1031-33. (pdf)
Bao XK, Liu NF, Qu JY, Wang XL, An B, Wen LY, and Song S (2010), The phylogenetic position and speciation dynamics of the genus Perdix (Phasianidae, Galliformes), Mol. Phylogenet. Evol. 56, 840-847. (abstract)
Birks SM, and Edwards SV (2002), A phylogeny of the megapodes (Aves: Megapodidae) based on nuclear and mitochondrial DNA sequences, Mol. Phylogenet. Evol. 23, 408-421. (abstract)
Bonilla AJ, Braun EL, and Kimball RT (2010), Comparative molecular evolution and phylogenetic utility of 3’-UTRs and introns in Galliformes, Mol. Phylogenet. Evol. 56, 536-542. (abstract)
Bowie RCK, Cohen C, and Crowe TM (2013), Ptilopachinae: a new subfamily of the Odontophoridae (Aves: Galliformes), Zootaxa 3670, 97-98. (abstract)
Cai T, Fjeldsa J, Wu Y, Shao S, Shen Y, Quan Q, Li X, Song X, Qu Y, Qiao G, and Lei F (2017), What makes the Sino-Himalayan mountains the major diversity hotspots for pheasants?, J. Biogeogr. 45, 640-651. (abstract)
Chen Y (2014), On the historical biogeography of global Galliformes: ancestral range and diversification patterns, Avian Res. 5, e:3. (pdf)
Chen D, Liu Y, Davison GWH, Dong L, Chang J, Gao S, Li SH, and Zhang Z (2015), Revival of the genus Tropicoperdix Blyth 1859 (Phasianidae, Aves) using multilocus sequence data, Zool. J. Linn. Soc. 175, 429-438. (abstract)
Chen D, Braun EL, Forthman M, Kimball RT, and Zhang Z (2018), A simple strategy for recovering ultraconserved elements, exons, and introns from low coverage shotgun sequencing of museum specimens: placement of the partridge genus Tropicoperdix within the galliformes, Mol. Phylogenet. Evol. 129, 304-314. (abstract)
Chen D, Liu Y, Davison G, Yong DL, Gao S, Hu J, Li SH, and Zhang Z (2020), Disentangling the evolutionary history and biogeography of hill partridges (Phasianidae, Arborophila) from low coverage shotgun sequences, Mol. Phylogenet. Evol. 151, e:106895. (abstract)
Chen D, Hosner PA, Dittmann DL, O‘Neill JP, Birks SM, Braun EL, and Kimball RT (2021), Divergence time estimation of Galliformes based on the best gene shopping scheme of ultraconserved elements, BMC Ecol. Evol. 21, e:209. (free pdf)
Cohen C, Wakeling JL, Mandiwana-Neudani TG, Sande E, Dranzoa C, Crowe TM, and Bowie RCK (2012), Phylogenetic affinities of evolutionary enigmatic African Galliformes: the Stone Partridge Ptilopachus petrosus and Nahan's Francolin Francolinus nahani, and support for their sister relationship with New World quails, Ibis 154, 768-780. (abstract)
Cox WA, Kimball RT, and Braun EL (2007), Phylogenetic position of the New World quail (Odontophoridae): eight nuclear loci and three mitochondrial regions contradict morphology and the Sibley-Ahlquist tapestry, Auk 124, 71-84. (free pdf)
Crowe TM, Bowie RCK, Bloomer P, Mandiwana TG, Hedderson TAJ, Randi E, Pereira SL, and Wakeling J (2006a), Phylogenetics, biogeography and classification of, and character evolution in, gamebirds (Aves: Galliformes): effects of character exclusion, data partitioning and missing data, Cladistics 22, 495-532. (pdf)
Crowe TM, Bloomer P, Randi E, Lucchini V, Kimball RT, Braun EL, and Groth JG (2006b), Supra-generic cladistics of landfowl (Order Galliformes), Acta Zool. Sinica 52, 358-361. (pdf)
Crowe TM, Mandiwana-Neudani TG, Donsker DB, Bowie RCK, and Little RM (2020), Resolving nomenclatural ‘confusion’ vis-à-vis Latham’s Francolin (Francolinus/ Peliperdix/Afrocolinus lathami) and the ‘Red-tailed’ francolins (Francolinus/Ortygornis/Peliperdix spp.), Ostrich 91, 1-3. (abstract)
Dey P, Ray SD, Kochiganti VHS, Pukazhenthi BS, Koepfli KP, and Singh RP (2024), Mitogenomic insights into the evolution, divergence time, and ancestral ranges of Coturnix quails, Genes 15, e:742. (free pdf)
Dimcheff DE, Drovetsky SV, and Mindell DP (2002), Phylogeny of Tetraoninae and other galliform birds using mitochondrial 12S and ND2 genes, Mol. Phylogenet. Evol. 24, 203-215. (abstract)
Ding L, Liao J, and Lin N (2019), The uplift of the Qinghai-Tibet Plateau and glacial oscillations triggered the diversification of Tetraogallus (Galliformes, Phasianidae), Ecol. Evol. 10, 1722-36. (pdf)
Drovestki SV (2002), Molecular phylogeny of grouse: individual and combined performance of W-linked, autosomal, and mitochondrial loci, Syst. Biol. 51, 930-945. (abstract)
Drovestki SV (2003), Plio-Pleistcene climatic oscilations, Holarctic biogeography and speciation in an avian subfamily, J. Biogeogr. 30, 1173-81. (abstract)
Forcina G, Panayides P, Guerrini M, Nardi F, Gupta BK, Mori E, Al-Sheikhli OF, Mansoori J, Khaliq I, Rank DN, Parasharya BM, Khan AA, Hadjigerou P, and Barbanera F (2012), Molecular evolution of the Asian francolins (Francolinus, Galliformes): a modern reappraisal of a classic study in speciation, Mol. Phylogenet. Evol. 65, 523-534. (abstract)
Frank-Hoeflich K, Silveira LF, Estudillo-Lopez J, Garcia-Koch AM, Ongay-Larios L, and Pinero D (2007), Increased taxon and character sampling reveals novel intergeneric relationships in the Cracidae, J. Zool. Syst. Evol. Res. 45, 242-254. (abstract)
Gao H, Liu Z, Sun Y, Zhao C, Wang J, and Teng L (2019), The complete mitochondrial genome of Helan Mountain chukar Alectoris chukar potanini (Galliformes: Phasianidae), Mitochondrial DNA Part B 4, 2443-44. (pdf)
Grau ET, Pereira SL, Silveira LF, Höfling E, and Wayntal A (2005), Molecular phylogenetics and biogeography of Neotropical piping guans (Aves: Galliformes): Pipile Bonaparte, 1856 is synonym of Aburria Reichenbach, 1853, Mol. Phylogenet. Evol. 35, 637-645. (abstract)
Guan X, Rao X, Song G, and Wang D (2022), The evolution of courtship displays in Galliformes, Avian Res. 13, e:100008. (free reading)
Gutierrez RJ, Barrowclough GF, and Groth JG (2000), A classification of the grouse (Aves: Tetraoninae) based on mitochondrial DNA sequences, Wildl. Biol. 6, 205-211. (free pdf)
Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han KL, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, and Yuri T (2008), A phylogenetic study of birds reveals their evolutionary history, Science 320, 1763-67. (abstract)
Harris RB, Birks SM, and Leach AD (2014), Incubator birds: biogeographical origins and evolution of underground nesting in megapodes (Galliformes: Megapodiidae), J. Biogeogr. 41, 2045-56. (pdf)
He L, Dai B, Zeng B, Zhang X, Chen B, Yue B, and Lee J (2009), The complete mitochondrial genome of the Sichuan Hill Partridge (Arborophila rufipectus) and a phylogenetic analysis with related species, Gene 435, 23-28. (abstract)
Hosner PA, Braun EL, and Kimball RT (2015), Land connectivity changes and global cooling shaped the colonization history and diversification of New World quail (Aves: Galliformes: Odontophoridae). J. Biogeogr. 42, 1883-95. (abstract)
Hosner PA, Faircloth BC, Glenn TC, Braun EL, and Kimball RT (2016a), Avoiding missing data biases in phylogenomic inference: an empirical study in the landfowl (Aves: Galliformes), Mol. Biol. Evol. 33, 1110-25. (free pdf)
Hosner PA, Braun EL, and Kimball RT (2016b), Rapid and recent diversification of curassows, guans, and chachalacas (Galliformes: Cracidae) out of Mesoamerica: phylogeny inferred from mitochondrial, intron, and ultraconserved element sequences, Mol. Phylogenet. Evol. 102, 320-330. (abstract)
Hosner PA, Tobias JA, Braun EL, and Kimball RT (2017), How do seemingly non-vagile clades accomplish trans-marine dispersal? Trait and dispersal evolution in the landfowl (Aves: Galliformes), Proc. R. Soc. B. 284, e:20170210. (pdf)
Hosner PA, Owens HL, Braun EL, and Kimball RT (2020), Phylogeny and diversification of the gallopheasants (Aves: Galliformes): Testing roles of sexual selection and environmental niche divergence, Zool. Scr. 49, 549-562. (abstract)
Jaiswal SK, Gupta A, Saxena R, Prasoodanan VPK, Sharma AK, Mittal P, Roy A, Shafer ABA, Vijay N, and Sharma VK (2018), Genome sequence of peacock reveals the peculiar case of a glittering bird, Front. Genet. 9, e:392. (pdf)
Jetz W, Thomas GH, Joy JB, Hartmann K, and Mooers AO (2012), The global diversity of birds in time and space, Nature 491, 444-448. (abstract)
Jiang F, Miao Y, Liang W, Ye H, Liu H, and Liu B (2010), The complete mitochondrial genomes of the whistling duck (Dendrocygna javanica) and black swan (Cygnus atratus): dating evolutionary divergence in Galloanserae, Mol. Biol. Rep. 37, 3001-15. (abstract)
Jiang L, Wang G, Peng R, Peng Q, and Zou F (2014), Phylogenetic and molecular dating analysis of Taiwan Blue Pheasant (Lophura swinhoii), Gene 539, 21-29. (abstract)
Kabasakal B, Doğru H, Erdoğan A, and Kaya S (2025), Transcontinental evolutionary dynamics and phylogeography of Alectoris (Aves: Galliformes): Identifying refuges, dispersal corridors, and cryptic diversity in the Palearctic region, Avian Res. e:100262. (free reading)
Kaiser VB, van Tuinen M, and Ellegren H (2007), Insertion events of CR1 retrotransposable elements elucidate the phylogenetic branching order in galliform birds, Mol. Biol. Evol. 24, 338-347. (free pdf)
Kan XZ, Yang JK, Li XF, Chen L, Lei ZP, Wang M, Qian CJ, Gao H, and Yang ZY (2010), Phylogeny of major lineages of galliform birds (Aves: Galliformes) based on complete mitochondrial genomes, Genet. Mol. Res. 9, 1625-33. (pdf)
Kimball RT, Braun EL, and Ligon JD (1997), Resolution of the phylogenetic position of the Congo peafowl, Afropavo congensis: a biogeographic and evolutionary enigma. Proc. R. Soc. Lond. B 264, 1517-23. (pdf)
Kimball RT, Braun EL, Zwartjes PW, Crowe TM, and Ligon JD (1999), A molecular phylogeny of the pheasants and partridges suggests that these lineages are not monophyletic. Mol. Phylogenet. Evol. 11, 38-45. (abstract)
Kimball RT, Braun EL, Ligon DJ, Lucchini V, and Randi E (2002), A molecular phylogeny of the peacock-pheasants (Galliformes Polyplectron spp.) indicates loss and reduction of ornamental traits and display behaviours, Biol. J. Linn. Soc. 73, 187-198. (abstract)
Kimball RT, and Braun EL (2008), A multigene phylogeny of Galliformes supports a single origin of erectile ability in non-feathered facial traits. J. Avian Biol. 39, 438-445. (pdf)
Kimball RT, St. Mary CM, and Braun EL (2011), A macroevolutionary perspective on multiple sexual traits in the Phasianidae (Galliformes). Int. J. Evol. Biol. 2011, e:423938. (pdf)
Kimball RT, and Braun EL (2014), Does more sequence data improve estimates of galliform phylogeny? Analyses of a rapid radiation using a complete data matrix. PeerJ 2, e:361. (pdf)
Kimball RT, Hosner PA, and Braun EL (2021a), A phylogenomic supermatrix of Galliformes (landfowl) reveals biased branch lengths. Mol. Phylogenet. Evol. 158, e:107091. (abstract)
Kimball RT, Guido M, Hosner PA, and Braun EL (2021b), When good mitochondria go bad: cyto-nuclear discordance in landfowl (Aves: Galliformes), Gene, 2021, e:145841. (abstract)
Kozma R, Rödin-Mörch P, and Höglund J (2017), Genomic regions of speciation and adaptation among three species of grouse. Sci. Rep. 9, e:812. (pdf)
Kriegs JO, Matzke A, Churakov G, Kuritzin A, Mayr G, Brosius J, and Schmitz J (2007), Waves of genomic hitchhikers shed light on the evolution of gamebirds (Aves: Galliformes), BMC Evol. Biol. 7, e:190. (pdf)
Ksepka DT (2009), Broken gears in the avian molecular clock: new phylogenetic analyses support stem galliform status for Gallinuloides wyomingensis and rallid affinities for Amitabha urbsinterdictensis, Cladistics 25, 173-197. (pdf)
Kuhl H, Frankl-Vilches C, Bakker A, Mayr G, Nikolaus G, Boerno ST, Klages S, Timmermann B, and Gahr M (2021), An unbiased molecular approach using 3'UTRs resolves the avian family-level tree of life, Mol. Biol. Evol. 38, 108-127. (free pdf)
Kumar S, Stecher G, Suleski M, and Hedges SB (2017), TimeTree: a resource for timelines, timetrees, and divergence times, Mol. Biol. Evol. 34, 1812-19. (free pdf)
Lee CY, Hsieh PH, Chiang LM, Chattopadhyay A, Li KY, Lee YF, Lu TP, Lai LC, Lin EC, Lee H, Ding ST, Tsai MH, Chen CY, and Chuang EY (2018), Whole-genome de novo sequencing reveals unique genes that contributed to the adaptive evolution of the Mikado pheasant, GigaScience 7, 1-14. (free pdf)
Lewis DC, Metallinos-Katzaras E, and Grivetti LE (1987), Coturnism: human poisoning by European Migratory Quail, J. Cult. Geogr. 7, 51-65. (abstract)
Li X, Huang Y, and Lei F (2015), Comparative mitochondrial genomics and phylogenetic relationships of the Crossoptilon species (Phasianidae, Galliformes), BMC Genomics 16, e:42. (pdf)
Liu F, Ma L, Yang C, Tu F, Xu Y, Ran J, Yue B, and Zhang X (2014), Taxonomic status of Tetraophasis obscurus and Tetraophasis szechenyii (Aves: Galliformes: Phasianidae) based on the complete mitochondrial genome, Zoolog. Sci. 31, 160-167. (abstract)
Liu Z, He L, Yuan H, Yue B, and Li J (2012), CR1 retroposons provide a new insight into the phylogeny of Phasianidae species (Aves: Galliformes), Gene 502, 125-132. (abstract)
Lucchini V, Höglund J, Klaus S, Swenson J, and Randi E (2001), Historical biogeography and a mitochondrial DNA phylogeny of grouse and ptarmigan. Mol. Phylogenet. Evol. 20, 149-162. (abstract)
Mandiwana-Neudani TG, Bowie RCK, Hausberger M, Henry L, and Crowe TM (2014), Taxonomic and phylogenetic utility of variation in advertising calls of francolins and spurfowls (Galliformes: Phasianidae). African Zool. 49, 54-82. (abstract)
Mandiwana-Neudani TG, Little RM, Crowe TM, and Bowie RCK (2019a), Taxonomy, phylogeny and biogeography of African spurfowls: Galliformes, Phasianidae, Coturnicini: Pternistis spp. Ostrich 90, 145-172. (abstract)
Mandiwana-Neudani TG, Little RM, Crowe TM, and Bowie RCK (2019b), Taxonomy, phylogeny and biogeography of ‘true’ francolins: Galliformes, Phasianidae, Phasianinae, Gallini: Francolinus, Ortygornis, Afrocolinus gen. nov., Peliperdix and Scleroptila spp., Ostrich 90, 191-221. (abstract)
Mayr G (2006), New specimens of the early Eocene stem group galliform Paraortygoides (Gallinuloididae), with comments on the evolution of a crop in the stem lineage of Galliformes, J. Ornithol. 147, 31-37. (abstract)
Mayr G (2016), "Avian Evolution: The Fossil Record of Birds and its Paleobiological Significance", Chapter 7, Wiley-Blackwell. (link)
Mayr G, and Weidig I (2004), The Early Eocene bird Gallinuloides wyomingensis - a stem group representative of Galliformes, Acta Palaeontol. Pol. 49, 211-217. (pdf)
Meiklejohn KA, Danielson MJ, Faircloth BC, Glenn TC, Braun EL, and Kimball RT (2014), Incongruence among different mitochondrial regions: a case study using complete mitogenomes. Mol. Phylogenet. Evol. 78, 314-323. (abstract)
Meiklejohn KA, Faircloth BC, Glenn TC, Kimball RT, and Braun EL (2016), Analysis of rapid evolutionary radiation using ultraconserved elements: evidence for a bias in some multispecies coalescent methods. Syst. Biol. 65, 612-627. (abstract)
Méndez-Camacho K, Leon-Alvarado O, and Miranda-Esquivel DR (2021), Biogeographic evidence supports the Old Amazon hypothesis for the formation of the Amazonian fluvial system, PeerJ 9, e:12533. (free pdf)
Pereira SL, Baker AJ, and Wajntal A (2002), Combined nuclear and mitochondrial DNA sequences resolve generic relationships within the Cracidae (Galliformes, Aves), Syst. Biol. 51, 946-958. (pdf)
Pereira SL, and Baker AJ (2004), Vicariant speciation of curassows (Aves, Cracidae): a hypothesis based on mitochondrial DNA phylogeny, Auk 123, 682-694. (pdf)
Pereira SL, and Baker AJ (2006), A molecular timescale for galliform birds accounting for uncertainty in time estimates and heterogeneity of rates of DNA substitutions across lineages and sites, Mol. Phylogenet. Evol. 38, 499-509. (abstract)
Pereira SL, and Baker AJ (2009), Waterfowl and gamefowl (Galloanserae), in "Timetree of Life", (Hedges, S.B., and S. Kumar, eds.), Oxford University Press, pp. 415-418. (pdf)
Pereira SL, Baker AJ, and Wajntal A (2009), Did increased taxon and character sampling really reveal novel intergeneric relationships in the Cracidae (Aves: Galliformes)?, J. Zool. Syst. Evol. Res. 47, 103-104. (abstract)
Persons NW, Hosner PA, Meiklejohn KA, Braun EL, and Kimball RT (2016), Sorting out relationships among the grouse and ptarmigan using intron, mitochondrial, and ultra-conserved element sequences, Mol. Phylogenet. Evol. 98, 123-132. (abstract)
Prum RO, Berv JS, Dornburg A, Field DJ, Townsend JP, Lemmon EM, and Lemmon AR (2015), A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing, Nature 526, 569-573. (abstract)
Randi E, Lucchini V, Armijo-Prewitt T, Kimball RT, Braun EL, and Ligon JD (2000), Mitochondrial DNA phylogeny and speciation in the tragopans, Auk 117, 1003-15. (pdf)
Román-Palacios C (2023), The ‘phruta R’ Package and ‘salphycon’ shiny app: increasing access, reproducibility, and transparency in phylogenetic analyses, bioRxiv (pdf)
Salter JF, Hosner PA, Tsai WLE, McCormack JE, Braun EL, Kimball RT, Brumfield RT, and Faircloth BC (2022), Historical specimens and the limits of subspecies phylogenomics in the New World quails (Odontophoridae), Mol. Phylogenet. Evol. 175, e:107559. (abstract)
Sangster G, Gregory SMS, and Dickinson EC (2023), Ithaginini, a new family-group name for the Blood Pheasant Ithaginis cruentus and resurrection of Lerwini for the Snow Partridge Lerwa lerwa (Phasianidae), Avian Syst. 2, 1-7. (pdf)
Seabrook-Davison M, Huynen L, Lambert DM, and Brunton DH (2009), Ancient DNA resolves identitiy and phylogeny of New Zealand´s extinct and living quail, PLOS ONE 4, e:6400. (pdf)
Shen YY, Liang L, Sun YB, Yue BS, Yang XJ, Murphy RW, and Zhang YP (2010), A mitogenomic perspective on the ancient, rapid radiation in the Galliformes with an emphasis on the Phasianidae, BMC Evol. Biol. 10, e:132. (pdf)
Shen YY, Dai K, Cao X, Murphy RW, Shen XJ, and Zhang YP (2014), The updated phylogenies of the Phasianidae based on combined data of nuclear and mitochondrial DNA, PLOS ONE 9, e:95786. (pdf)
Stein RW,
Brown
JW, and Mayr G (2015), A molecular genetic time scale demonstrates Cretaceous origins and multiple diversification rate shifts within the order Galliformes (Aves), Mol.
Phylogent. Evol. 92, 155-164.
(abstract)
Sun K, Meiklejohn KA, Faircloth BC, Glenn TC, Braun EL, and Kimball RT (2014), The evolution of peafowl and other taxa with ocelli (eyespots): a phylogenomic approach, Proc. R. Soc. B 281, e:20140823. (pdf)
Sveinsdottir M, and Magnusson KP (2014), Complete mitochondrial genome and phylogenetic analysis of willow ptarmigan (Lagopus lagopus) and rock ptarmigan (Lagopus muta) (Galliformes: Phasianidae: Tetraoninae), Mitochondrial DNA Part B 2, 400-402. (pdf)
Tiley GP, Pandey A, Kimball RT, Braun EL, and Burleigh JG (2020), Whole genome phylogeny of Gallus: introgression and data-type effects. Avian Res. 11 , e:7. (pdf)
Tomek T, Bochenski ZM, Wertz K, and Swidnicka E (2014), A new genus and species of a galliform bird from the Oligocene of Poland, Palaeontol. Electron. 17, e:38A. (pdf)
Wang, N., R.T. Kimball, E.L. Braun, B. Liang, and Z. Zhang (2013), Assessing phylogenetic relationships among Galliformes: a multigene phylogeny with extended taxon sampling, PLOS ONE 8, e:64312. (pdf)
Wang N, Kimball RT, Braun EL, Liang B, and Zhang Z (2017a), Ancestral range reconstruction of Galliformes: the effects of topology and taxon sampling in Phasianidae, J. Biogeogr. 44, 122-135. (abstract)
Wang N, Hosner PA, Liang B, Braun EL, Liang B, and Kimball RT (2017b), Historical relationships of three enigmatic phasianid genera (Aves: Galliformes) inferred using phylogenomic and mitogenomic data, Mol. Phylogenet. Evol. 109, 217-225. (abstract)
Wang P, Liu Y, Chang Y, Wang N, and Zhang Z (2017c), The role of niche divergence and geographic arrangement in the speciation of Eared Pheasants (Crossoptilon, Hodgson 1938), Mol. Phylogenet. Evol. 113, 1-8. (abstract)
Williford D, DeYoung RW, and Brennan LA (2017), Molecular ecology of New World Quails: messages for managers, National Quail Symposium Proceedings 8, article 20. (pdf)
Yan C, Mou B, Meng Y, Tu F, Fan Z, Price M, Yue B, and Zhang X (2017), A novel mitochondrial genome of Arborophila and new insight into Arborophila evolutionary history, PLOS ONE 12, e:0181649. (pdf)
Zhan XJ, and Zhang ZW (2005), Molecular phylogeny of avian genus Syrmaticus based on the mitochondrial cytochrome b gene and control region, Zool. Sci. 22, 427-435. (abstract)
Zhao S, Ma Y, Wang G, Li H, Liu X, Yu J, Yue B, and Zou F (2012), Molecular phylogeny of major lineages of the avian family Phasianidae inferred from complete mitochondrial genome sequences, J. Nat. Hist. 46, 757-767. (abstract)
Zhou C, Zheng S, Jiang X, Liang W, Price M, Fan Z, Meng Y, and Yue B (2018), First complete genome sequence in Arborophila and comparative genomics reveals the evolutionary adaptation of Hainan Partridge (Arborophila ardens), Avian Res. 9, e:45. (pdf)
Zhou C, Liu Y, Qiao L, Liu Y, Yang N, Meng Y, and Yue B (2020), The draft genome of the blood pheasant (Ithaginis cruentus): phylogeny and high-altitude adaptation, Ecol. Evol. 10, 11440-52. (pdf)
Zhou C, Zheng X, Feng K, Peng K, Zhang Y, Zhao G, Meng Y, Zhang L, Yue B, and Wu Y (2023), The draft genome of the Tibetan partridge (Perdix hodgsoniae) provides insights into its phylogenetic position and high-altitude adaptation, J. Hered. 114, 275-188. (abstract)
Photos
Cracidae
Numididae
Phasianidae















